6BRE
 
 | | Hen Egg-White Lysozyme cocrystallized with Cadmium sulphate using CuKalpha source. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Sakon, J, Caviness, P, Jack, T, Pickens, J.B, Reed, P, Ruth, C. | | Deposit date: | 2017-11-30 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Hen Egg-White Lysozyme (HEWL) cocrystallized with Cadmium sulphate.
To Be Published
|
|
6I82
 
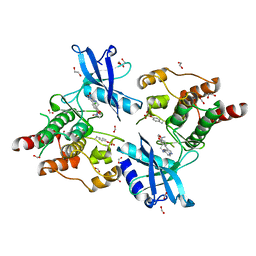 | | Crystal structure of partially phosphorylated RET V804M tyrosine kinase domain complexed with PDD00018412 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Burschowsky, D, Seewooruthun, C, Bayliss, R, Carr, M.D, Echalier, A, Jordan, A.M. | | Deposit date: | 2018-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Optimization of wt-RET/KDR-Selective Inhibitors of RETV804MKinase.
Acs Med.Chem.Lett., 11, 2020
|
|
6I83
 
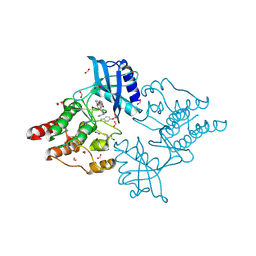 | | Crystal structure of phosphorylated RET V804M tyrosine kinase domain complexed with PDD00018366 | | Descriptor: | 4-[5-(pyridin-3-ylmethylamino)pyrazolo[1,5-a]pyrimidin-3-yl]benzamide, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Burschowsky, D, Seewooruthun, C, Bayliss, R, Carr, M.D, Echalier, A, Jordan, A.M. | | Deposit date: | 2018-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and Optimization of wt-RET/KDR-Selective Inhibitors of RETV804MKinase.
Acs Med.Chem.Lett., 11, 2020
|
|
6SS4
 
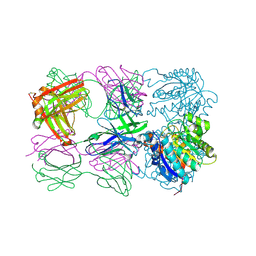 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021181 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021181 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS2
 
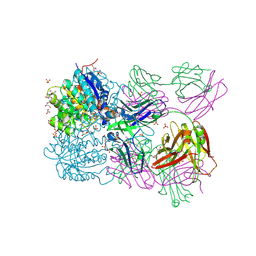 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0021158 | | Descriptor: | Arginase-2, mitochondrial, Fab C0021158 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS5
 
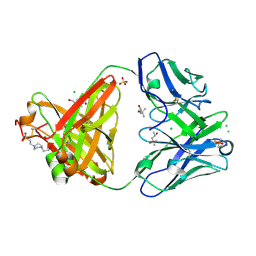 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0020187 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Extensive sequence and structural evolution of Arginase 2 inhibitory antibodies enabled by an unbiased approach to affinity maturation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TUL
 
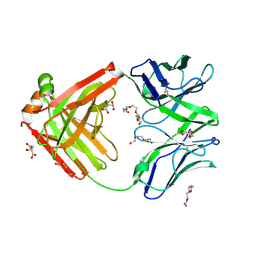 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021177 | | Descriptor: | D-MALATE, Fab C0021177 heavy chain (IgG1), Fab C0021177 light chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2020-01-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS0
 
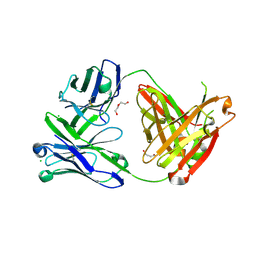 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021181 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fab C0021181 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SRX
 
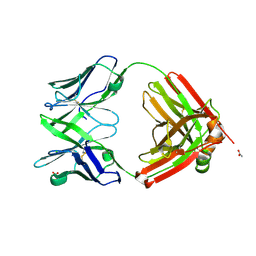 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021158 | | Descriptor: | ACETATE ION, CHLORIDE ION, Fab C0021158 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SS6
 
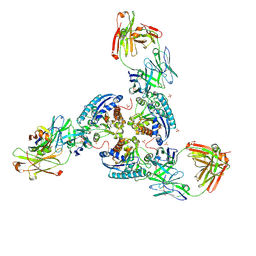 | | Structure of arginase-2 in complex with the inhibitory human antigen-binding fragment Fab C0020187 | | Descriptor: | Arginase-2, mitochondrial, Fab C0020187 heavy chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Extensive sequence and structural evolution of Arginase 2 inhibitory antibodies enabled by an unbiased approach to affinity maturation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SRV
 
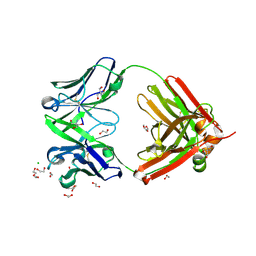 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021144 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
5IKU
 
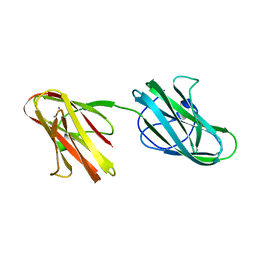 | | Crystal structure of the Hathewaya histolytica ColG tandem collagen-binding domain s3as3b in the presence of calcium at 1.9 Angstrom resolution | | Descriptor: | CALCIUM ION, Collagenase | | Authors: | Janowska, K, Bauer, R, Roeser, R, Sakon, J, Matsushita, O. | | Deposit date: | 2016-03-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ca2+-induced orientation of tandem collagen binding domains from clostridial collagenase ColG permits two opposing functions of collagen fibril formation and retardation.
Febs J., 285, 2018
|
|
