6U7N
 
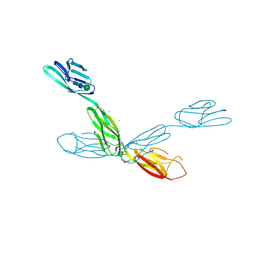 | | Crystal structure of neurotrimin (NTM) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Neurotrimin, ... | | Authors: | Machius, M, Venkannagari, H, Misra, A, Rudenko, G, Rush, S. | | Deposit date: | 2019-09-03 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.321 Å) | | Cite: | Highly Conserved Molecular Features in IgLONs Contrast Their Distinct Structural and Biological Outcomes.
J.Mol.Biol., 432, 2020
|
|
6E36
 
 | |
6E3F
 
 | |
8FK4
 
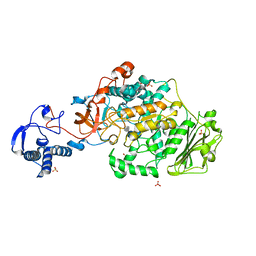 | | Structure of the catalytic domain of Streptococcus mutans GtfB complexed to acarbose in orthorhombic space group P21212 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, Glucosyltransferase-I, ... | | Authors: | Schormann, N, Deivanayagam, C. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The catalytic domains of Streptococcus mutans glucosyltransferases: a structural analysis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8FJ9
 
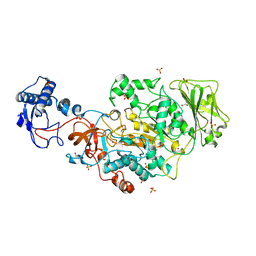 | | Structure of the catalytic domain of Streptococcus mutans GtfB in tetragonal space group P4322 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schormann, N, Deivanayagam, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic domains of Streptococcus mutans glucosyltransferases: a structural analysis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8FJC
 
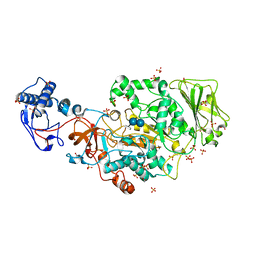 | | Structure of the catalytic domain of Streptococcus mutans GtfB complexed to acarbose in tetragonal space group P4322 | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Schormann, N, Deivanayagam, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic domains of Streptococcus mutans glucosyltransferases: a structural analysis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8FN5
 
 | |
8FKL
 
 | |
6CAM
 
 | |
4EIJ
 
 | |
6SRR
 
 | |
6SQU
 
 | | Crystal structure of human SHIP2 catalytic domain in complex with 1,2,4 Dimer | | Descriptor: | 5,5'-(ethane-1,2-diylbis(oxy))bis(benzene-5,4,2,1,-tetrayl)hexakisphosphate, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | | Authors: | Whitfield, H, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2019-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Allosteric Site on SHIP2 Identified Through Fluorescent Ligand Screening and Crystallography: A Potential New Target for Intervention.
J.Med.Chem., 64, 2021
|
|
7L0O
 
 | |
