4W64
 
 | | Hcp1 protein from Acinetobacter baumannii AB0057 | | Descriptor: | SULFATE ION, Type VI secretion system effector, Hcp1 family | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2014-08-20 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Hcp from Acinetobacter baumannii: A Component of the Type VI Secretion System.
Plos One, 10, 2015
|
|
4WVV
 
 | |
4WVW
 
 | | Chicken Galectin-8 N-terminal domain complexed with 3'-sialyl-lactose | | Descriptor: | DI(HYDROXYETHYL)ETHER, Galectin, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2014-11-07 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Combining Crystallography and Hydrogen-Deuterium Exchange to Study Galectin-Ligand Complexes.
Chemistry, 21, 2015
|
|
5JP5
 
 | | Crystal structure of rat Galectin 5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Galectin-5 | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2016-05-03 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Characterization of Rat Galectin-5, an N-Tailed Monomeric Proto-Type-like Galectin.
Biomolecules, 11, 2021
|
|
6Y4R
 
 | | Cytoplasmic domain of TssL from Acinetobacter baumannii | | Descriptor: | DotU family type IV/VI secretion system protein | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2020-02-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural Characterization of TssL from Acinetobacter baumannii: a Key Component of the Type VI Secretion System.
J.Bacteriol., 202, 2020
|
|
5NLD
 
 | | Chicken GRIFIN (crystallisation pH: 7.5) | | Descriptor: | Galectin, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2017-04-04 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Chicken GRIFIN: Structural characterization in crystals and in solution.
Biochimie, 146, 2018
|
|
5NM1
 
 | | Chicken GRIFIN (crystallisation pH: 6.2) | | Descriptor: | Galectin, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2017-04-05 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Chicken GRIFIN: Structural characterization in crystals and in solution.
Biochimie, 146, 2018
|
|
5NLH
 
 | | Chicken GRIFIN (crystallisation pH: 8.5) | | Descriptor: | Galectin, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2017-04-04 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Chicken GRIFIN: Structural characterization in crystals and in solution.
Biochimie, 146, 2018
|
|
5NLZ
 
 | | GRIFIN (Crystallisation pH: 4.2) | | Descriptor: | Galectin | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2017-04-05 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Chicken GRIFIN: Structural characterization in crystals and in solution.
Biochimie, 146, 2018
|
|
5NM6
 
 | |
5NLE
 
 | | Chicken GRIFIN (crystallisation pH: 8.0) | | Descriptor: | Galectin, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2017-04-04 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Chicken GRIFIN: Structural characterization in crystals and in solution.
Biochimie, 146, 2018
|
|
5NMJ
 
 | | Chicken GRIFIN (crystallisation pH: 6.5) | | Descriptor: | Galectin, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2017-04-06 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Chicken GRIFIN: Structural characterization in crystals and in solution.
Biochimie, 146, 2018
|
|
7P8H
 
 | | chicken GRIFIN bound to blood group tetrasaccharide B (type 1) | | Descriptor: | Galectin, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2021-07-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structural Characterization of Rat Galectin-5, an N-Tailed Monomeric Proto-Type-like Galectin.
Biomolecules, 11, 2021
|
|
2YMZ
 
 | | Crystal structure of chicken Galectin 2 | | Descriptor: | GALECTIN 2, SULFATE ION, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Fernandez, I.S, Ruiz, F.M, Solis, D, Gabius, H.-J, Romero, A. | | Deposit date: | 2012-10-11 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fine-Tuning of Prototype Chicken Galectins: Structure of Cg-2 and Structure-Activity Correlations
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3LSE
 
 | | N-Domain of human adhesion/growth-regulatory galectin-9 in complex with lactose | | Descriptor: | Galectin-9, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2010-02-12 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | N-domain of human adhesion/growth-regulatory galectin-9: preference for distinct conformers and non-sialylated N-glycans and detection of ligand-induced structural changes in crystal and solution.
Int.J.Biochem.Cell Biol., 42, 2010
|
|
3LSD
 
 | | N-Domain of human adhesion/growth-regulatory galectin-9 | | Descriptor: | Galectin-9 | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2010-02-12 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | N-domain of human adhesion/growth-regulatory galectin-9: preference for distinct conformers and non-sialylated N-glycans and detection of ligand-induced structural changes in crystal and solution.
Int.J.Biochem.Cell Biol., 42, 2010
|
|
5JPG
 
 | | Rat Galectin 5 with lactose | | Descriptor: | Galectin-5, SODIUM ION, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Romero, A, Ruiz, F.M. | | Deposit date: | 2016-05-03 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Characterization of Rat Galectin-5, an N-Tailed Monomeric Proto-Type-like Galectin.
Biomolecules, 11, 2021
|
|
7ON3
 
 | | SaFtsZ complexed with GDP (soak 10 mM EGTA) | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OJD
 
 | | SaFtsZ(D46A) complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OMP
 
 | | SaFtsZ complexed with GDPPCP and Mg2+ | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, MAGNESIUM ION, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OJA
 
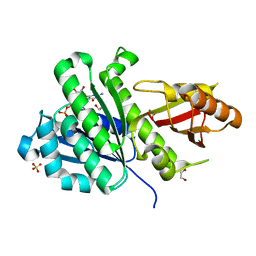 | | SaFtsZ(D210N) complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OHH
 
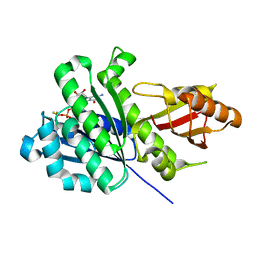 | | SaFtsZ complexed with GDP and BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-11 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OJC
 
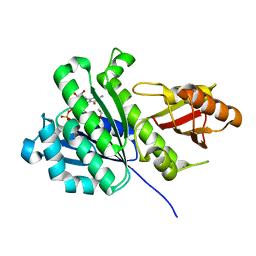 | | SaFtsZ(Q48A) complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7OMQ
 
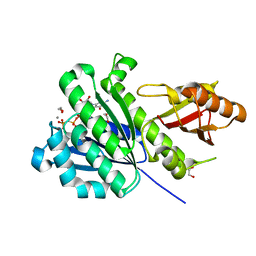 | | SaFtsZ complexed with GDPPCP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, MANGANESE (II) ION, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
7ON4
 
 | | SaFtsZ complexed with GDP (co-crystalization with 1mM EDTA) | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Tornero, C, Ruiz, F.M, Andreu, J.M. | | Deposit date: | 2021-05-25 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics.
Plos Biol., 20, 2022
|
|
