3L32
 
 | |
1VYI
 
 | | Structure of the c-terminal domain of the polymerase cofactor of rabies virus: insights in function and evolution. | | Descriptor: | GLYCEROL, RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Mavrakis, M, McCarthy, A.A, Roche, S, Blondel, D, Ruigrok, R.W.H. | | Deposit date: | 2004-04-30 | | Release date: | 2004-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Function of the C-Terminal Domain of the Polymerase Cofactor of Rabies Virus
J.Mol.Biol., 343, 2004
|
|
6XTE
 
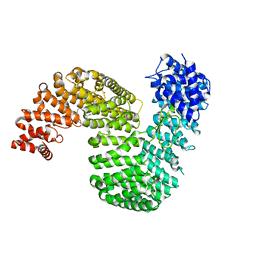 | | Human karyopherin RanBP5 (isoform-1) | | Descriptor: | Antipain, CHLORIDE ION, Importin-5, ... | | Authors: | Swale, C, McCarthy, A.A, Ruigrok, R.W.H, Crepin, T. | | Deposit date: | 2020-01-16 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | X-ray Structure of the Human Karyopherin RanBP5, an Essential Factor for Influenza Polymerase Nuclear Trafficking.
J.Mol.Biol., 432, 2020
|
|
6XU2
 
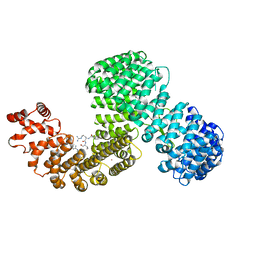 | | Human karyopherin RanBP5 (isoform-3) | | Descriptor: | Antipain, Importin-5, NICKEL (II) ION | | Authors: | Swale, C, McCarthy, A.A, Berger, I, Bieniossek, C, Delmas, B, Ruigrok, R.W.H, Crepin, T. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.834 Å) | | Cite: | X-ray Structure of the Human Karyopherin RanBP5, an Essential Factor for Influenza Polymerase Nuclear Trafficking.
J.Mol.Biol., 432, 2020
|
|
1NSC
 
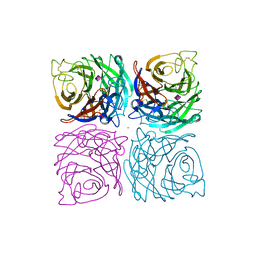 | | INFLUENZA B VIRUS NEURAMINIDASE CAN SYNTHESIZE ITS OWN INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Burmeister, W.P, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 1993-05-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influenza B virus neuraminidase can synthesize its own inhibitor.
Structure, 1, 1993
|
|
1NSD
 
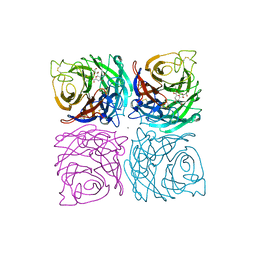 | | INFLUENZA B VIRUS NEURAMINIDASE CAN SYNTHESIZE ITS OWN INHIBITOR | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Burmeister, W.P, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 1993-05-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influenza B virus neuraminidase can synthesize its own inhibitor.
Structure, 1, 1993
|
|
1T3E
 
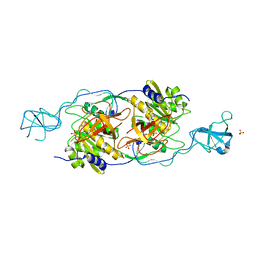 | | Structural basis of dynamic glycine receptor clustering | | Descriptor: | 49-mer fragment of Glycine receptor beta chain, Gephyrin, SULFATE ION | | Authors: | Sola, M, Bavro, V.N, Timmins, J, Franz, T, Ricard-Blum, S, Schoehn, G, Ruigrok, R.W.H, Paarmann, I, Saiyed, T, O'Sullivan, G.A. | | Deposit date: | 2004-04-26 | | Release date: | 2004-07-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis of dynamic glycine receptor clustering by gephyrin
Embo J., 23, 2004
|
|
8PZQ
 
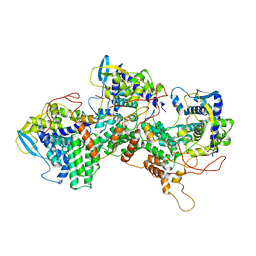 | | Model for focused reconstruction of influenza A RNP-like particle | | Descriptor: | Nucleoprotein, RNA (5'P-(UC)6-FAM3') | | Authors: | Chenavier, F, Estrozi, L.F, Zarkadas, E, Ruigrok, R.W.H, Schoehn, G, Ballandras-Colas, A, Crepin, T. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Cryo-EM structure of influenza helical nucleocapsid reveals NP-NP and NP-RNA interactions as a model for the genome encapsidation.
Sci Adv, 9, 2023
|
|
8PZP
 
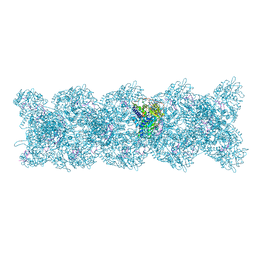 | | Model for influenza A virus helical ribonucleoprotein-like structure | | Descriptor: | Nucleoprotein, RNA (5'P-(UC)6-FAM3') | | Authors: | Chenavier, F, Estrozi, L.F, Zarkadas, E, Ruigrok, R.W.H, Schoehn, G, Ballandras-Colas, A, Crepin, T. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-27 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Cryo-EM structure of influenza helical nucleocapsid reveals NP-NP and NP-RNA interactions as a model for the genome encapsidation.
Sci Adv, 9, 2023
|
|
4UFT
 
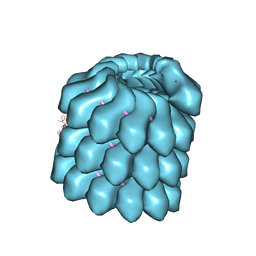 | | Structure of the helical Measles virus nucleocapsid | | Descriptor: | 5'-R(*CP*CP*CP*CP*CP*CP)-3', NUCLEOPROTEIN | | Authors: | Gutsche, I, Desfosses, A, Effantin, G, Ling, W.L, Haupt, M, Ruigrok, R.W.H, Sachse, C, Schoehn, G. | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-Atomic Cryo-Em Structure of the Helical Measles Virus Nucleocapsid.
Science, 348, 2015
|
|
4V4U
 
 | | The quasi-atomic model of Human Adenovirus type 5 capsid | | Descriptor: | HEXON PROTEIN, N-TERMINAL PEPTIDE OF FIBER PROTEIN, PENTON PROTEIN | | Authors: | Fabry, C.M.S, Rosa-Calatrava, M, Conway, J.F, Zubieta, C, Cusack, S, Ruigrok, R.W.H, Schoehn, G. | | Deposit date: | 2005-03-03 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | A Quasi-Atomic Model of Human Adenovirus Type 5 Capsid.
Embo J., 24, 2005
|
|
2GTT
 
 | | Crystal structure of the rabies virus nucleoprotein-RNA complex | | Descriptor: | Nucleoprotein, PHOSPHATE ION, RNA (99-MER) | | Authors: | Albertini, A.A.V, Wernimont, A.K, Muziol, T, Ravelli, R.B.G, Weissenhorn, W, Ruigrok, R.W.H. | | Deposit date: | 2006-04-28 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal Structure of the Rabies Virus Nucleoprotein-RNA Complex
Science, 313, 2006
|
|
8B8A
 
 | | Multimerization domain of borna disease virus 1 phosphoprotein | | Descriptor: | Phosphoprotein | | Authors: | Tarbouriech, N, Legrand, P, Bourhis, J.M, Chenavier, F, Freslon, L, Kawasaki, J, Horie, M, Tomonaga, K, Bachiri, K, Coyaud, E, Gonzalez-Dunia, D, Ruigrok, R.W.H, Crepin, T. | | Deposit date: | 2022-10-04 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses, 14, 2022
|
|
6Q8J
 
 | | Nterminal domain of human SMU1 in complex with LSP641 | | Descriptor: | 1-~{tert}-butyl-3-[6-(3,5-dimethoxyphenyl)-2-[[1-[1-(phenylmethyl)piperidin-4-yl]-1,2,3-triazol-4-yl]methylamino]pyrido[2,3-d]pyrimidin-7-yl]urea, WD40 repeat-containing protein SMU1 | | Authors: | Tengo, L, Le Corre, L, Fournier, G, Ashraf, U, Busca, P, Rameix-Welti, M.-A, Gravier-Pelletier, C, Ruigrok, R.W.H, Jacob, Y, Vidalain, P.-O, Pietrancosta, N, Naffakh, N, McCarthy, A.A, Crepin, T. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Destabilization of the human RED-SMU1 splicing complex as a basis for host-directed antiinfluenza strategy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6Q8I
 
 | | Nterminal domain of human SMU1 in complex with human REDmid | | Descriptor: | Protein Red, WD40 repeat-containing protein SMU1 | | Authors: | Tengo, L, Le Corre, L, Fournier, G, Ashraf, U, Busca, P, Rameix-Welti, M.-A, Gravier-Pelletier, C, Ruigrok, R.W.H, Jacob, Y, Vidalain, P.-O, Pietrancosta, N, Naffakh, N, McCarthy, A.A, Crepin, T. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Destabilization of the human RED-SMU1 splicing complex as a basis for host-directed antiinfluenza strategy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2C9F
 
 | | THE QUASI-ATOMIC MODEL OF THE ADENOVIRUS TYPE 3 PENTON DODECAHEDRON | | Descriptor: | FIBER, PENTON PROTEIN | | Authors: | Fuschiotti, P, Schoehn, G, Fender, P, Fabry, C.M.S, Hewat, E.A, Chroboczek, J, Ruigrok, R.W.H, Conway, J.F. | | Deposit date: | 2005-12-12 | | Release date: | 2006-03-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16.5 Å) | | Cite: | Structure of the Dodecahedral Penton Particle from Human Adenovirus Type 3.
J.Mol.Biol., 356, 2006
|
|
2C9G
 
 | | THE QUASI-ATOMIC MODEL OF THE ADENOVIRUS TYPE 3 PENTON BASE DODECAHEDRON | | Descriptor: | PENTON PROTEIN | | Authors: | Fuschiotti, P, Schoehn, G, Fender, P, Fabry, C.M.S, Hewat, E.A, Chroboczek, J, Ruigrok, R.W.H, Conway, J.F. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Structure of the Dodecahedral Penton Particle from Human Adenovirus Type 3.
J.Mol.Biol., 356, 2006
|
|
6Q8F
 
 | | Nterminal domain of human SMU1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, WD40 repeat-containing protein SMU1 | | Authors: | Tengo, L, Le Corre, L, Fournier, G, Ashraf, U, Busca, P, Rameix-Welti, M.-A, Gravier-Pelletier, C, Ruigrok, R.W.H, Jacob, Y, Vidalain, P.-O, Pietrancosta, N, Naffakh, N, McCarthy, A.A, Crepin, T. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Destabilization of the human RED-SMU1 splicing complex as a basis for host-directed antiinfluenza strategy.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2JDQ
 
 | | C-terminal domain of influenza A virus polymerase PB2 subunit in complex with human importin alpha5 | | Descriptor: | IMPORTIN ALPHA-1 SUBUNIT, POLYMERASE BASIC PROTEIN 2 | | Authors: | Tarendeau, F, Guilligay, D, Mas, P, Boulo, S, Baudin, F, Ruigrok, R.W.H, Hart, D.J, Cusack, S. | | Deposit date: | 2007-01-11 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Nuclear Import Function of the C- Terminal Domain of Influenza Virus Polymerase Pb2 Subunit
Nat.Struct.Mol.Biol., 14, 2007
|
|
1NSB
 
 | |
2VQZ
 
 | | Structure of the cap-binding domain of influenza virus polymerase subunit PB2 with bound m7GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, POLYMERASE BASIC PROTEIN 2 | | Authors: | Guilligay, D, Tarendeau, F, Resa-Infante, P, Coloma, R, Crepin, T, Sehr, P, Lewis, J, Ruigrok, R.W.H, Ortin, J, Hart, D.J, Cusack, S. | | Deposit date: | 2008-03-21 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structural Basis for CAP Binding by Influenza Virus Polymerase Subunit Pb2.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VTW
 
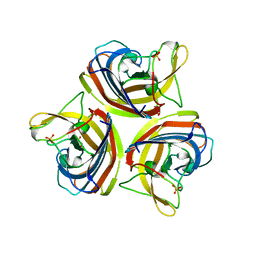 | | Structure of the C-terminal head domain of the fowl adenovirus type 1 short fibre | | Descriptor: | FIBER PROTEIN 2, GLYCEROL, SULFATE ION | | Authors: | ElBakkouri, M, Seiradake, E, Cusack, S, Ruigrok, R.W.H, Schoehn, G. | | Deposit date: | 2008-05-16 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the C-Terminal Head Domain of the Fowl Adenovirus Type 1 Short Fibre.
Virology, 378, 2008
|
|
2W69
 
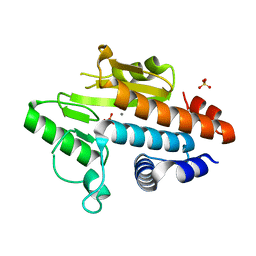 | | Influenza polymerase fragment | | Descriptor: | MANGANESE (II) ION, POLYMERASE ACIDIC PROTEIN, SULFATE ION | | Authors: | Dias, A, Bouvier, D, Crepin, T, McCarthy, A.A, Hart, D.J, Baudin, F, Cusack, S, Ruigrok, R.W.H. | | Deposit date: | 2008-12-17 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The CAP-Snatching Endonuclease of Influenza Virus Polymerase Resides in the Pa Subunit
Nature, 458, 2009
|
|
1O6E
 
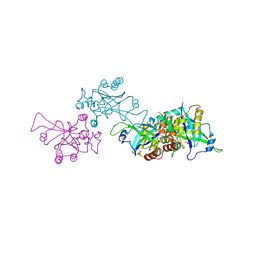 | | Epstein-Barr virus protease | | Descriptor: | CAPSID PROTEIN P40, PHOSPHORYLISOPROPANE | | Authors: | Buisson, M, Hernandez, J, Lascoux, D, Schoehn, G, Forest, E, Arlaud, G, Seigneurin, J, Ruigrok, R.W.H, Burmeister, W.P. | | Deposit date: | 2002-09-13 | | Release date: | 2002-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the Epstein-Barr Virus Protease Shows Rearrangement of the Processed C Terminus
J.Mol.Biol., 324, 2002
|
|
9GAQ
 
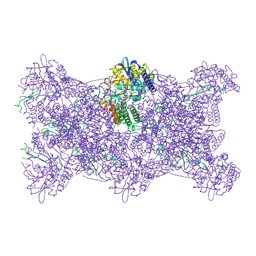 | | CryoEM structure of influenza A RNP-like particle double-stranded assembled with a 14-mer RNA. | | Descriptor: | Nucleoprotein, RNA (5'-P(UC)7-FAM3') | | Authors: | Chenavier, F, Ruigrok, R.W.H, Schoehn, G, Ballandras-Colas, A, Crepin, T. | | Deposit date: | 2024-07-29 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Influenza a virus antiparallel helical nucleocapsid-like pseudo-atomic structure.
Nucleic Acids Res., 53, 2025
|
|
