3TNX
 
 | | Structure of the precursor of a thermostable variant of papain at 2.6 Angstroem resolution | | 分子名称: | CHLORIDE ION, Papain | | 著者 | Roy, S, Choudhury, D, Dattagupta, J.K, Biswas, S. | | 登録日 | 2011-09-02 | | 公開日 | 2012-09-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | The structure of a thermostable mutant of pro-papain reveals its activation mechanism
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1VEQ
 
 | | Mycobacterium smegmatis Dps Hexagonal form | | 分子名称: | FE (III) ION, starvation-induced DNA protecting protein | | 著者 | Roy, S, Gupta, S, Das, S, Sekar, K, Chatterji, D, Vijayan, M. | | 登録日 | 2004-04-03 | | 公開日 | 2004-06-29 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.98 Å) | | 主引用文献 | X-ray analysis of Mycobacterium smegmatis Dps and a comparative study involving other Dps and Dps-like molecules
J.Mol.Biol., 339, 2004
|
|
1VEL
 
 | | Mycobacterium smegmatis Dps tetragonal form | | 分子名称: | CADMIUM ION, SODIUM ION, SULFATE ION, ... | | 著者 | Roy, S, Gupta, S, Das, S, Sekar, K, Chatterji, D, Vijayan, M. | | 登録日 | 2004-04-01 | | 公開日 | 2004-06-29 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | X-ray analysis of Mycobacterium smegmatis Dps and a comparative study involving other Dps and Dps-like molecules
J.Mol.Biol., 339, 2004
|
|
1VEI
 
 | | Mycobacterium smegmatis Dps | | 分子名称: | FE (III) ION, SULFATE ION, starvation-induced DNA protecting protein | | 著者 | Roy, S, Gupta, S, Das, S, Sekar, K, Chatterji, D, Vijayan, M. | | 登録日 | 2004-03-31 | | 公開日 | 2004-06-29 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | X-ray Analysis of Mycobacterium smegmatis Dps and a Comparative Study Involving Other Dps and Dps-like Molecules
J.Mol.Biol., 339, 2004
|
|
4S1E
 
 | |
4S1J
 
 | |
3BT4
 
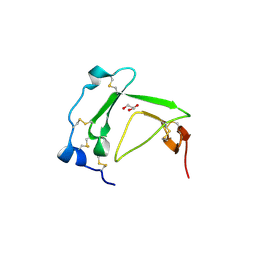 | | Crystal Structure Analysis of AmFPI-1, fungal protease inhibitor from Antheraea mylitta | | 分子名称: | Fungal protease inhibitor-1, GLYCEROL | | 著者 | Roy, S, Aravind, P, Madhurantakam, C, Ghosh, A.K, Sankarananarayanan, R, Das, A.K. | | 登録日 | 2007-12-27 | | 公開日 | 2008-12-30 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of a fungal protease inhibitor from Antheraea mylitta
J.Struct.Biol., 166, 2009
|
|
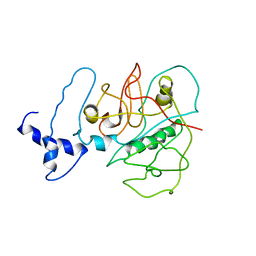
3USV
 
 | |
7M1S
 
 | |
2YW6
 
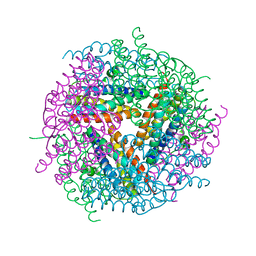 | | Structural studies of N terminal deletion mutant of Dps from Mycobacterium smegmatis | | 分子名称: | DNA protection during starvation protein | | 著者 | Roy, S, Saraswathi, R, Gupta, S, Sekar, K, Chatterji, D, Vijayan, M. | | 登録日 | 2007-04-19 | | 公開日 | 2007-07-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Role of N and C-terminal Tails in DNA Binding and Assembly in Dps: Structural Studies of Mycobacterium smegmatis Dps Deletion Mutants
J.Mol.Biol., 370, 2007
|
|
2YW7
 
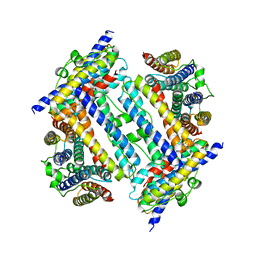 | | Crystal structure of C-terminal deletion mutant of Mycobacterium smegmatis Dps | | 分子名称: | Starvation-induced DNA protecting protein | | 著者 | Roy, S, Saraswathi, R, Gupta, S, Sekar, K, Chatterji, D, Vijayan, M. | | 登録日 | 2007-04-19 | | 公開日 | 2007-07-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Role of N and C-terminal Tails in DNA Binding and Assembly in Dps: Structural Studies of Mycobacterium smegmatis Dps Deletion Mutants
J.Mol.Biol., 370, 2007
|
|
6VKJ
 
 | |
3LGF
 
 | |
3LGL
 
 | |
3LH0
 
 | |
2Z90
 
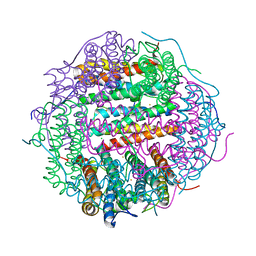 | | Crystal Structure of the Second Dps from Mycobacterium smegmatis | | 分子名称: | CHLORIDE ION, FE (II) ION, MAGNESIUM ION, ... | | 著者 | Roy, S, Saraswathi, R, Chatterji, D, Vijayan, M. | | 登録日 | 2007-09-13 | | 公開日 | 2008-04-22 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural studies on the second Mycobacterium smegmatis Dps: invariant and variable features of structure, assembly and function.
J.Mol.Biol., 375, 2008
|
|
4ONO
 
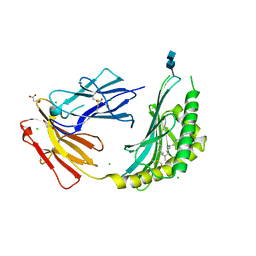 | | CD1c in complex with PM (phosphomycoketide) | | 分子名称: | (4R,8S,16S,20R)-4,8,12,16,20-pentamethylheptacosyl dihydrogen phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin/T-cell surface glycoprotein CD1c/T-cell surface glycoprotein CD1b chimeric protein, ... | | 著者 | Roy, S, Adams, E.J. | | 登録日 | 2014-01-28 | | 公開日 | 2014-10-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.705 Å) | | 主引用文献 | Molecular basis of mycobacterial lipid antigen presentation by CD1c and its recognition by alpha beta T cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4ONH
 
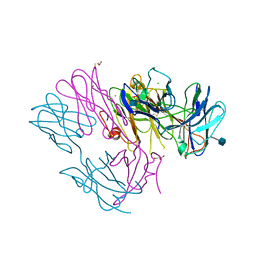 | | Crystal Structure of DN6 TCR | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | 著者 | Roy, S, Adams, E.J. | | 登録日 | 2014-01-28 | | 公開日 | 2014-10-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.008 Å) | | 主引用文献 | Molecular basis of mycobacterial lipid antigen presentation by CD1c and its recognition by alpha beta T cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7EB1
 
 | | Solution NMR structure of the RRM domain of RNA binding protein RBM3 from homo sapiens | | 分子名称: | RNA-binding protein 3 | | 著者 | Boral, S, Roy, S, Basak, A.J, Maiti, S, Lee, W, De, S. | | 登録日 | 2021-03-08 | | 公開日 | 2021-12-08 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and dynamic studies of the human RNA binding protein RBM3 reveals the molecular basis of its oligomerization and RNA recognition.
Febs J., 289, 2022
|
|
7P7I
 
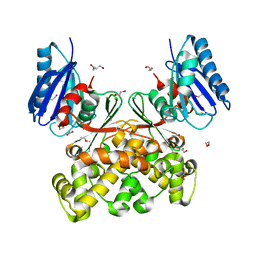 | | Native structure of N-acetylglucosamine kinase from Plesiomonas shigelloides | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | 登録日 | 2021-07-19 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7PA1
 
 | | Structure of N-acetylglucosamine kinase from Plesiomonas shigelloides in complex with AMP-PNP in the absence of N-acetylglucoseamine substrate | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | 著者 | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | 登録日 | 2021-07-28 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9Y
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | 著者 | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | 登録日 | 2021-07-28 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9L
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine-6-phosphate | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-6-O-phosphono-beta-D-glucopyranose, ... | | 著者 | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | 登録日 | 2021-07-27 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P7W
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine and ADP | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | 登録日 | 2021-07-20 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9P
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine and AMP-PNP inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | 登録日 | 2021-07-27 | | 公開日 | 2022-08-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
