4PLP
 
 | |
1G28
 
 | |
4TVB
 
 | |
1JNU
 
 | |
3KXE
 
 | |
2R13
 
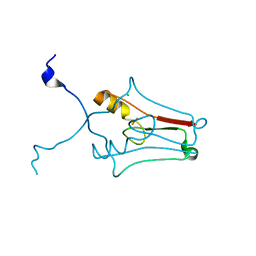 | | Crystal structure of human mitoNEET reveals a novel [2Fe-2S] cluster coordination | | Descriptor: | CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, Zinc finger CDGSH domain-containing protein 1 | | Authors: | Hou, X, Liu, R, Ross, S, Smart, E.J, Zhu, H, Gong, W. | | Deposit date: | 2007-08-22 | | Release date: | 2007-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic studies of human MitoNEET
J.Biol.Chem., 282, 2007
|
|
6ZQT
 
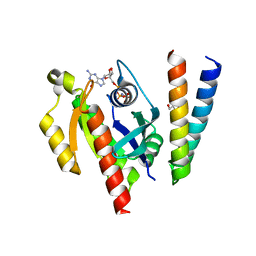 | | Crystal structure of the RLIP76 Ral binding domain mutant (E427H/Q433L/K440R) in complex with RalB-GMPPNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Hurd, C, Brear, P, Revell, J, Ross, S, Mott, H, Owen, D. | | Deposit date: | 2020-07-10 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Affinity maturation of the RLIP76 Ral binding domain to inform the design of stapled peptides targeting the Ral GTPases.
J.Biol.Chem., 296, 2020
|
|
6ZRN
 
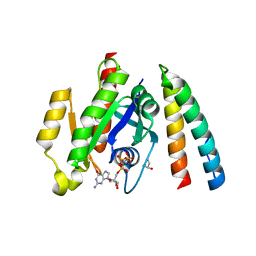 | | Crystal structure of the RLIP76 Ral binding domain mutant (E427S/L429M/Q433L/K440R) in complex with RalB-GMPPNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Hurd, C, Brear, P, Revell, J, Ross, S, Mott, H, Owen, D. | | Deposit date: | 2020-07-13 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Affinity maturation of the RLIP76 Ral binding domain to inform the design of stapled peptides targeting the Ral GTPases.
J.Biol.Chem., 296, 2020
|
|
5DYN
 
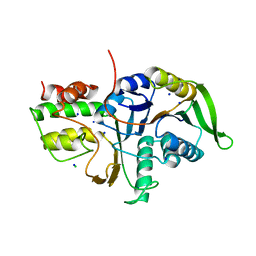 | | B. fragilis cysteine protease | | Descriptor: | CHLORIDE ION, Putative peptidase, SODIUM ION | | Authors: | Choi, V.M, Herrou, J, Hecht, A.L, Turner, J.R, Crosson, S, Bubeck Wardenburg, J. | | Deposit date: | 2015-09-24 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Activation of Bacteroides fragilis toxin by a novel bacterial protease contributes to anaerobic sepsis in mice.
Nat. Med., 22, 2016
|
|
3UNW
 
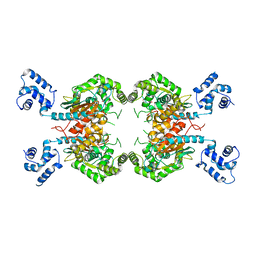 | | Crystal Structure of Human GAC in Complex with Glutamate | | Descriptor: | GLUTAMIC ACID, Glutaminase kidney isoform, mitochondrial | | Authors: | DeLaBarre, B, Gross, S, Cheng, F, Gao, Y, Jha, A, Jiang, F, Song, J.J, Wie, W, Hurov, J. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Full-length human glutaminase in complex with an allosteric inhibitor.
Biochemistry, 50, 2011
|
|
3UO9
 
 | | Crystal Structure of Human GAC in Complex with Glutamate and BPTES | | Descriptor: | GLYCEROL, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | DeLaBarre, B, Gross, S, Cheng, F, Gao, Y, Jha, A, Jiang, F, Song, J.J, Wei, W, Hurov, J.B. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Full-length human glutaminase in complex with an allosteric inhibitor.
Biochemistry, 50, 2011
|
|
6NTR
 
 | | Crystal Structure of Beta-barrel-like Protein of Domain of Unknown Function DUF1849 from Brucella abortus | | Descriptor: | 1,2-ETHANEDIOL, ATP/GTP-binding site-containing protein A, GLYCEROL | | Authors: | Kim, Y, Bigelow, L, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-01-30 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | BrucellaPeriplasmic Protein EipB Is a Molecular Determinant of Cell Envelope Integrity and Virulence.
J.Bacteriol., 201, 2019
|
|
3NAD
 
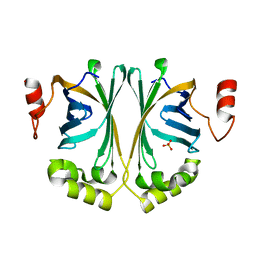 | | Crystal Structure of Phenolic Acid Decarboxylase from Bacillus pumilus UI-670 | | Descriptor: | Ferulate decarboxylase, SULFATE ION | | Authors: | Matte, A, Grosse, S, Bergeron, H, Abokitse, K, Lau, P.C.K. | | Deposit date: | 2010-06-01 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural analysis of Bacillus pumilus phenolic acid decarboxylase, a lipocalin-fold enzyme.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3N0R
 
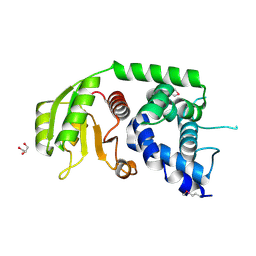 | |
5F4B
 
 | | Structure of B. abortus WrbA-related protein A (WrpA) | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, NAD(P)H dehydrogenase (quinone) | | Authors: | Herrou, J, Czyz, D, Willett, J.W, Kim, H.S, Chhor, G, Endres, M, Babnigg, G, Kim, Y, Joachimiak, A, Crosson, S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | WrpA Is an Atypical Flavodoxin Family Protein under Regulatory Control of the Brucella abortus General Stress Response System.
J.Bacteriol., 198, 2016
|
|
5F51
 
 | | Structure of B. abortus WrbA-related protein A (apo) | | Descriptor: | NAD(P)H dehydrogenase (quinone), SULFATE ION | | Authors: | Herrou, J, Czyz, D, Willett, J.W, Kim, H.S, Kim, Y, Crosson, S. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | WrpA Is an Atypical Flavodoxin Family Protein under Regulatory Control of the Brucella abortus General Stress Response System.
J.Bacteriol., 198, 2016
|
|
5UC0
 
 | | Crystal Structure of Beta-barrel-like, Uncharacterized Protein of COG5400 from Brucella abortus | | Descriptor: | CHLORIDE ION, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Kim, Y, Bigelow, L, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-12-21 | | Release date: | 2017-02-08 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Periplasmic protein EipA determines envelope stress resistance and virulence in Brucella abortus.
Mol. Microbiol., 2018
|
|
5TEU
 
 | | Brucella periplasmic binding protein YehZ | | Descriptor: | ACETATE ION, L(+)-TARTARIC ACID, SULFATE ION, ... | | Authors: | Herrou, J, Crosson, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Conserved ABC Transport System Regulated by the General Stress Response Pathways of Alpha- and Gammaproteobacteria.
J. Bacteriol., 199, 2017
|
|
5TF3
 
 | | Crystal Structure of Protein of Unknown Function YPO2564 from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, Putative membrane protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Anderson, W.F, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-23 | | Release date: | 2016-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Protein of Unknown Function YPO2564 from Yersinia pestis
To Be Published
|
|
5UVE
 
 | | Crystal Structure of the ABC Transporter Substrate-binding protein BAB1_0226 from Brucella abortus | | Descriptor: | CALCIUM ION, GLYCEROL, Substrate-binding region of ABC-type glycine betaine transport system | | Authors: | Kim, Y, Chhor, G, Endres, M, Hero, J, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-02-20 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Beta-barrel-like Protein of Unknown Function
To Be Published
|
|
4Q0C
 
 | | 3.1 A resolution crystal structure of the B. pertussis BvgS periplasmic domain | | Descriptor: | Virulence sensor protein BvgS | | Authors: | Dupre, E, Herrou, J, Lensink, M.F, Wintjens, R, Lebedev, A, Crosson, S, Villeret, V, Locht, C, Antoine, R, Jacob-Dubuisson, F. | | Deposit date: | 2014-04-01 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Virulence Regulation with Venus Flytrap Domains: Structure and Function of the Periplasmic Moiety of the Sensor-Kinase BvgS.
Plos Pathog., 11, 2015
|
|
4QPJ
 
 | | 2.7 Angstrom Structure of a Phosphotransferase in Complex with a Receiver Domain | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cell cycle response regulator CtrA, ... | | Authors: | Willett, J.W, Herrou, J, Crosson, S. | | Deposit date: | 2014-06-23 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.742 Å) | | Cite: | Structural asymmetry in a conserved signaling system that regulates division, replication, and virulence of an intracellular pathogen.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4QPK
 
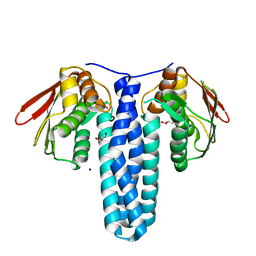 | | 1.7 Angstrom Structure of a Bacterial Phosphotransferase | | Descriptor: | GLYCEROL, PHOSPHATE ION, PHOSPHOTRANSFERASE, ... | | Authors: | Willett, J.W, Crosson, S, Herrou, J. | | Deposit date: | 2014-06-23 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural asymmetry in a conserved signaling system that regulates division, replication, and virulence of an intracellular pathogen.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4IRX
 
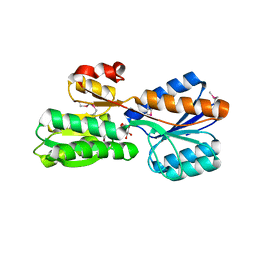 | | Crystal structure of Caulobacter myo-inositol binding protein bound to myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, Sugar ABC transporter, periplasmic sugar-binding protein | | Authors: | Herrou, J, Crosson, S. | | Deposit date: | 2013-01-15 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | myo-inositol and D-ribose ligand discrimination in an ABC periplasmic binding protein.
J.Bacteriol., 195, 2013
|
|
4N27
 
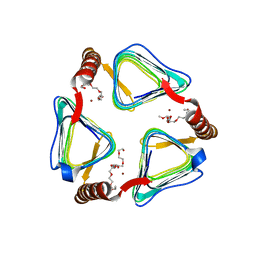 | | X-ray structure of Brucella abortus RicA | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Bacterial transferase hexapeptide repeat, ZINC ION | | Authors: | Herrou, J, Crosson, S. | | Deposit date: | 2013-10-04 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular Structure of the Brucella abortus Metalloprotein RicA, a Rab2-Binding Virulence Effector.
Biochemistry, 52, 2013
|
|
