7YXA
 
 | | XFEL crystal structure of the human sphingosine 1 phosphate receptor 5 in complex with ONO-5430608 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[6-(2-naphthalen-1-ylethoxy)-2,3,4,5-tetrahydro-1H-3-benzazepin-3-ium-3-yl]butanoic acid, ... | | Authors: | Lyapina, E, Marin, E, Gusach, A, Orekhov, P, Gerasimov, A, Luginina, A, Vakhrameev, D, Ergasheva, M, Kovaleva, M, Khusainov, G, Khorn, P, Shevtsov, M, Kovalev, K, Okhrimenko, I, Bukhdruker, S, Popov, P, Hu, H, Weierstall, U, Liu, W, Cho, Y, Gushchin, I, Rogachev, A, Bourenkov, G, Park, S, Park, G, Huyn, H.J, Park, J, Gordeliy, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for receptor selectivity and inverse agonism in S1P 5 receptors.
Nat Commun, 13, 2022
|
|
7BMH
 
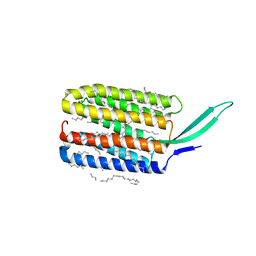 | | Crystal structure of a light-driven proton pump LR (Mac) from Leptosphaeria maculans | | Descriptor: | EICOSANE, OLEIC ACID, Opsin | | Authors: | Kovalev, K, Zabelskii, D, Dmitrieva, N, Volkov, O, Shevchenko, V, Astashkin, R, Zinovev, E, Gordeliy, V. | | Deposit date: | 2021-01-20 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based insights into evolution of rhodopsins.
Commun Biol, 4, 2021
|
|
6EYU
 
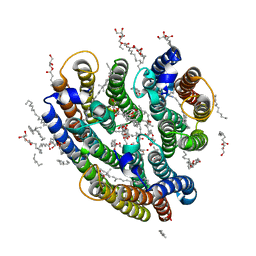 | | Crystal structure of the inward H(+) pump xenorhodopsin | | Descriptor: | Bacteriorhodopsin, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Shevchenko, V, Polovinkin, V, Mager, T, Gushchin, I, Melnikov, I, Borshchevskiy, V, Popov, A, Alekseev, A, Gordeliy, V. | | Deposit date: | 2017-11-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inward H(+) pump xenorhodopsin: Mechanism and alternative optogenetic approach.
Sci Adv, 3, 2017
|
|
6YT4
 
 | | Crystal structure of the N112A mutant of the light-driven sodium pump KR2 in the pentameric form, pH 8.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Maliar, N, Astashkin, R, Gordeliy, V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the N112A Mutant of the Light-Driven Sodium Pump KR2
Crystals, 2020
|
|
7AKW
 
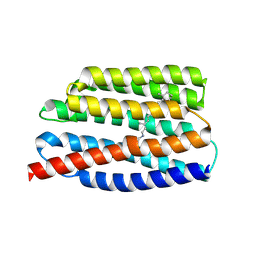 | | Crystal structure of the viral rhodopsins chimera O1O2 | | Descriptor: | EICOSANE, RETINAL, chimera of viral rhodopsins OLPVR1 and OLPVRII | | Authors: | Kovalev, K, Zabelskii, D, Alekseev, A, Astashkin, R, Gordeliy, V. | | Deposit date: | 2020-10-02 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Viral rhodopsins 1 are an unique family of light-gated cation channels.
Nat Commun, 11, 2020
|
|
7AKY
 
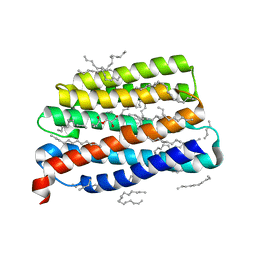 | | Crystal structure of the viral rhodopsin OLPVR1 in P21212 space group | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-hexadec-9-enoate, EICOSANE, viral rhodopsin OLPVR1 | | Authors: | Kovalev, K, Zabelskii, D, Alekseev, A, Astashkin, R, Gordeliy, V. | | Deposit date: | 2020-10-02 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Viral rhodopsins 1 are an unique family of light-gated cation channels.
Nat Commun, 11, 2020
|
|
7AKX
 
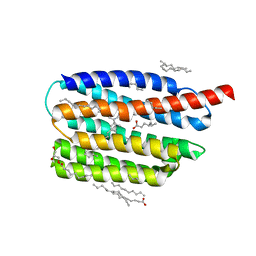 | | Crystal structure of the viral rhodopsin OLPVR1 in P1 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Zabelskii, D, Alekseev, A, Astashkin, R, Gordeliy, V. | | Deposit date: | 2020-10-02 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Viral rhodopsins 1 are an unique family of light-gated cation channels.
Nat Commun, 11, 2020
|
|
7PZY
 
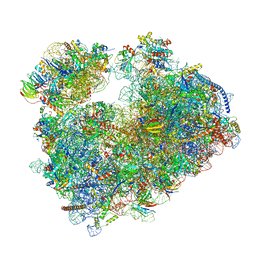 | | Structure of the vacant Candida albicans 80S ribosome | | Descriptor: | 1,4-DIAMINOBUTANE, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7Q0F
 
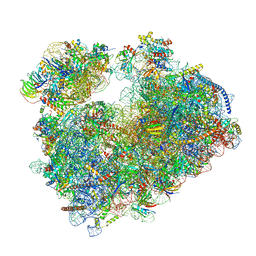 | | Structure of Candida albicans 80S ribosome in complex with phyllanthoside | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7Q0P
 
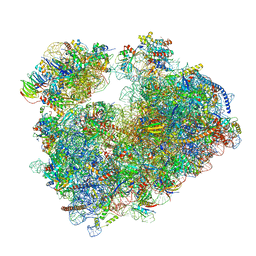 | | Structure of the Candida albicans 80S ribosome in complex with anisomycin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-15 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7Q08
 
 | | Structure of Candida albicans 80S ribosome in complex with cycloheximide | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7Q0R
 
 | | Structure of the Candida albicans 80S ribosome in complex with blasticidin s | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-16 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
6T0H
 
 | | Crystal structure of CYP124 in complex with 1-alpha-hydroxy-vitamin D3 | | Descriptor: | 1-alpha-hydroxy-vitamin D3, CHLORIDE ION, CYP124 in complex with inhibitor carbethoxyhexyl imidazole, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
6T0G
 
 | | Crystal structure of CYP124 in complex with vitamin D3 | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, MAGNESIUM ION, Methyl-branched lipid omega-hydroxylase, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
6T0K
 
 | | Crystal structure of CYP124 in complex with inhibitor carbethoxyhexyl imidazole | | Descriptor: | CHLORIDE ION, CYP124 in complex with inhibitor carbethoxyhexyl imidazole, GLYCEROL, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
6T0F
 
 | | Crystal structure of CYP124 in complex with cholest-4-en-3-one | | Descriptor: | (8ALPHA,9BETA)-CHOLEST-4-EN-3-ONE, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
6T0L
 
 | | Crystal structure of CYP124 in complex with inhibitor compound 5' | | Descriptor: | CHLORIDE ION, CYP124 in complex with inhibitor compound 5', DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bukhdruker, S, Marin, E, Varaksa, T, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Metabolic Fate of Human Immunoactive Sterols in Mycobacterium tuberculosis.
J.Mol.Biol., 433, 2021
|
|
6SU3
 
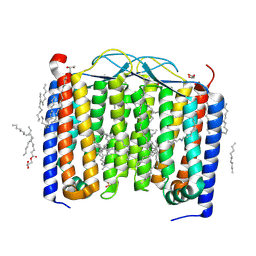 | | Crystal structure of the 48C12 heliorhodopsin in the violet form at pH 8.8 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 48C12 heliorhodopsin, EICOSANE, ... | | Authors: | Kovalev, K, Volkov, D, Astashkin, R, Alekseev, A, Gushchin, I, Gordeliy, V. | | Deposit date: | 2019-09-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structural insights into the heliorhodopsin family.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SU4
 
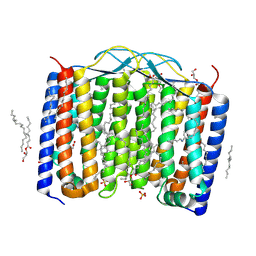 | | Crystal structure of the 48C12 heliorhodopsin in the blue form at pH 4.3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 48C12 heliorhodopsin, ACETATE ION, ... | | Authors: | Kovalev, K, Volkov, D, Astashkin, R, Alekseev, A, Gushchin, I, Gordeliy, V. | | Deposit date: | 2019-09-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structural insights into the heliorhodopsin family.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7ZB9
 
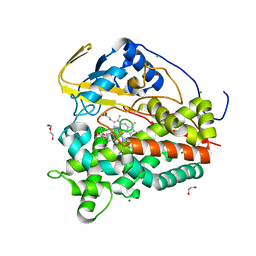 | | Crystal structure of CYP124 in complex with inhibitor carbethoxyhexyl imidazole in the absence of glycerol (NoCryo) | | Descriptor: | CHLORIDE ION, CYP124 in complex with inhibitor carbethoxyhexyl imidazole, MAGNESIUM ION, ... | | Authors: | Bukhdruker, S, Varaksa, T, Marin, E, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the effects of glycerol on ligand binding to cytochrome P450.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7ZMY
 
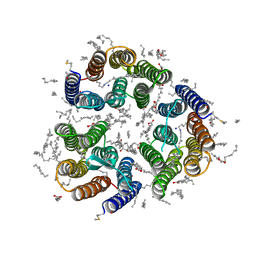 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the ground state at pH 8.2 in the presence of sodium at 100K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZN0
 
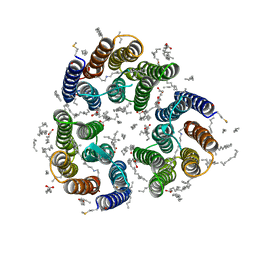 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 8.2 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZN3
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the L state at pH 8.2 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZN9
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 7.0 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZNB
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 5.2 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
