1IH5
 
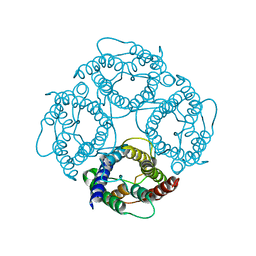 | | CRYSTAL STRUCTURE OF AQUAPORIN-1 | | Descriptor: | AQUAPORIN-1 | | Authors: | Ren, G, Reddy, V.S, Cheng, A, Melnyk, P, Mitra, A.K. | | Deposit date: | 2001-04-18 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.7 Å) | | Cite: | Visualization of a water-selective pore by electron crystallography in vitreous ice.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3DYR
 
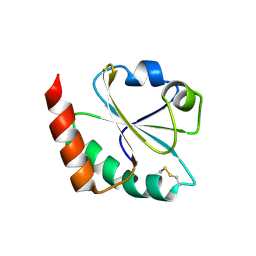 | |
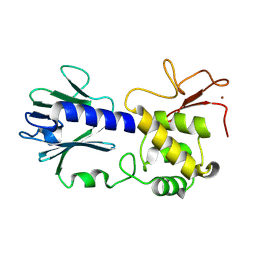
8TJG
 
 | |
8R3T
 
 | | Cofactor-free Tau 4R2N isoform | | Descriptor: | Microtubule-associated protein tau | | Authors: | Limorenko, G, Tatli, M, Kolla, R, Nazarov, S, Weil, M.T, Schondorf, D.C, Geist, D, Reinhardt, P, Ehrnhoefer, D.E, Stahlberg, H, Gasparini, L, Lashuel, H.A. | | Deposit date: | 2023-11-10 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Fully co-factor-free ClearTau platform produces seeding-competent Tau fibrils for reconstructing pathological Tau aggregates.
Nat Commun, 14, 2023
|
|
1HIO
 
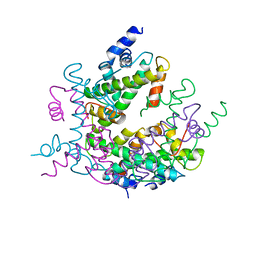 | | HISTONE OCTAMER (CHICKEN), CHROMOSOMAL PROTEIN, ALPHA CARBONS ONLY | | Descriptor: | HISTONE H2A, HISTONE H2B, HISTONE H3, ... | | Authors: | Arents, G, Moudrianakis, E.N. | | Deposit date: | 1991-09-19 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
2HIO
 
 | | HISTONE OCTAMER (CHICKEN), CHROMOSOMAL PROTEIN | | Descriptor: | PROTEIN (HISTONE H2A), PROTEIN (HISTONE H2B), PROTEIN (HISTONE H3), ... | | Authors: | Arents, G, Moudrianakis, E.N. | | Deposit date: | 1999-06-15 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
5KBZ
 
 | | Structure of the PksA Product Template domain in complex with a phosphopantetheine mimetic | | Descriptor: | (14R)-14-hydroxy-15,15-dimethyl-1-[5-({[(5-methyl-1,2-oxazol-3-yl)methyl]sulfanyl}methyl)-1,2-oxazol-3-yl]-4,9,13-trioxo-2-thia-5,8,12-triazahexadecan-16-yl dihydrogen phosphate, Noranthrone synthase | | Authors: | Tsai, S.C, Burkart, M.D, Townsend, C.A, Barajas, J.F, Shakya, G, Topper, C.L, Moreno, G, Jackson, D.R, Rivera, H, La Clair, J.J, Vagstad, A. | | Deposit date: | 2016-06-03 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Polyketide mimetics yield structural and mechanistic insights into product template domain function in nonreducing polyketide synthases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4QVR
 
 | | 2.3 Angstrom Crystal Structure of Hypothetical Protein FTT1539c from Francisella tularensis. | | Descriptor: | Uncharacterized hypothetical protein FTT_1539c | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Flores, K, Ren, G, Huntley, J.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-15 | | Release date: | 2014-07-30 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Crystal Structure of Hypothetical Protein FTT1539c from Francisella tularensis.
TO BE PUBLISHED
|
|
4ENZ
 
 | | Structure of human ceruloplasmin at 2.6 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Samygina, V.R, Sokolov, A.V, Bourenkov, G, Vasilyev, V.B, Bartunik, H. | | Deposit date: | 2012-04-13 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ceruloplasmin: macromolecular assemblies with iron-containing acute phase proteins.
Plos One, 8, 2013
|
|
1L8X
 
 | | Crystal Structure of Ferrochelatase from the Yeast, Saccharomyces cerevisiae, with Cobalt(II) as the Substrate Ion | | Descriptor: | COBALT (II) ION, Ferrochelatase | | Authors: | Karlberg, T, Lecerof, D, Gora, M, Silvegren, G, Labbe-Bois, R, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-03-22 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Metal Binding to Saccharomyces cerevisiae Ferrochelatase
Biochemistry, 41, 2002
|
|
4UWM
 
 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2014-08-12 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7YXA
 
 | | XFEL crystal structure of the human sphingosine 1 phosphate receptor 5 in complex with ONO-5430608 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[6-(2-naphthalen-1-ylethoxy)-2,3,4,5-tetrahydro-1H-3-benzazepin-3-ium-3-yl]butanoic acid, ... | | Authors: | Lyapina, E, Marin, E, Gusach, A, Orekhov, P, Gerasimov, A, Luginina, A, Vakhrameev, D, Ergasheva, M, Kovaleva, M, Khusainov, G, Khorn, P, Shevtsov, M, Kovalev, K, Okhrimenko, I, Bukhdruker, S, Popov, P, Hu, H, Weierstall, U, Liu, W, Cho, Y, Gushchin, I, Rogachev, A, Bourenkov, G, Park, S, Park, G, Huyn, H.J, Park, J, Gordeliy, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for receptor selectivity and inverse agonism in S1P 5 receptors.
Nat Commun, 13, 2022
|
|
7Z09
 
 | | Crystal structure of the ground state of bacteriorhodopsin at 1.05 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z0A
 
 | | Crystal structure of the ground state of bacteriorhodopsin at 1.22 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z0D
 
 | | Crystal structure of the L state of bacteriorhodopsin at 1.20 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z0C
 
 | | Crystal structure of the K state of bacteriorhodopsin at 1.53 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7Z0E
 
 | | Crystal structure of the M state of bacteriorhodopsin at 1.22 Angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6E,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene, 2,3-DI-PHYTANYL-GLYCEROL, ... | | Authors: | Borshchevskiy, V, Kovalev, K, Round, E, Efremov, R, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-02-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | True-atomic-resolution insights into the structure and functional role of linear chains and low-barrier hydrogen bonds in proteins.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6SV9
 
 | | Non-terahertz irradiated structure of bovine trypsin (even frames of crystal x40) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
6SVU
 
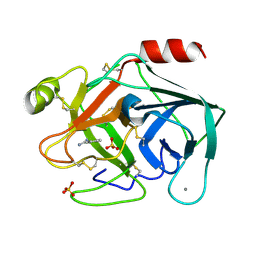 | | Reference structure of bovine trypsin (even frames of crystal x30) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
6SVZ
 
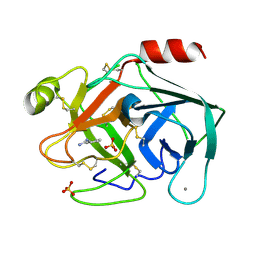 | | Reference structure of bovine trypsin (even frames of crystal x34) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
6SVB
 
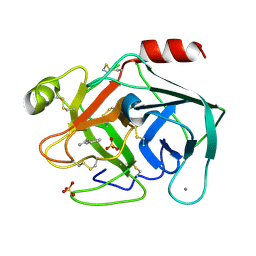 | | Terahertz irradiated structure of bovine trypsin (odd frames of crystal x40) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
6SVJ
 
 | | Terahertz irradiated structure of bovine trypsin (odd frames of crystal x42) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
6SVR
 
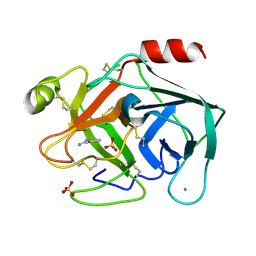 | | Reference structure of bovine trypsin (odd frames of crystal x28) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
6SV6
 
 | | Non-terahertz irradiated structure of bovine trypsin (even frames of crystal x38) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | Deposit date: | 2019-09-17 | | Release date: | 2020-01-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
6SVN
 
 | | Reference structure of bovine trypsin (even frames of crystal x28) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Ahlberg Gagner, V, Lundholm, I, Garcia-Bonete, M.J, Rodilla, H, Friedman, R, Zhaunerchyk, V, Bourenkov, G, Schneider, T, Stake, J, Katona, G. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-22 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Clustering of atomic displacement parameters in bovine trypsin reveals a distributed lattice of atoms with shared chemical properties.
Sci Rep, 9, 2019
|
|
