4B4P
 
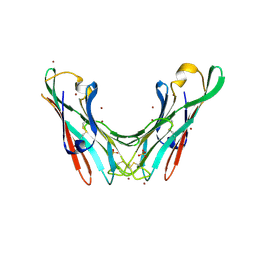 | | Crystal Structure of the lectin domain of F18 fimbrial adhesin FedF. | | Descriptor: | BROMIDE ION, F18 FIMBRIAL ADHESIN AC, SULFATE ION | | Authors: | Moonens, K, Bouckaert, J, Coddens, A, Tran, T, Panjikar, S, De Kerpel, M, Cox, E, Remaut, H, De Greve, H. | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-15 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insight in Histo-Blood Group Binding by the F18 Fimbrial Adhesin Fedf.
Mol.Microbiol., 86, 2012
|
|
4B4Q
 
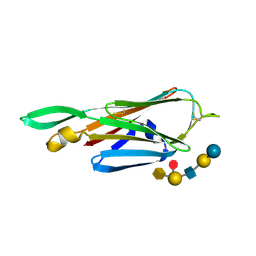 | | Crystal Structure of the lectin domain of F18 fimbrial adhesin FedF in complex with blood group A type 1 hexasaccharide | | Descriptor: | F18 FIMBRIAL ADHESIN AC, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Moonens, K, Bouckaert, J, Coddens, A, Tran, T, Panjikar, S, De Kerpel, M, Cox, E, Remaut, H, De Greve, H. | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight in Histo-Blood Group Binding by the F18 Fimbrial Adhesin Fedf.
Mol.Microbiol., 86, 2012
|
|
4B4R
 
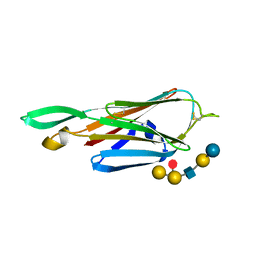 | | Crystal Structure of the lectin domain of F18 fimbrial adhesin FedF in complex with blood group B type 1 hexasaccharide | | Descriptor: | F18 FIMBRIAL ADHESIN AC, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Moonens, K, Bouckaert, J, Coddens, A, Tran, T, Panjikar, S, De Kerpel, M, Cox, E, Remaut, H, De Greve, H. | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insight in Histo-Blood Group Binding by the F18 Fimbrial Adhesin Fedf.
Mol.Microbiol., 86, 2012
|
|
3ZBI
 
 | | Fitting result in the O-layer of the subnanometer structure of the bacterial pKM101 type IV secretion system core complex digested with elastase | | Descriptor: | TRAF PROTEIN, TRAN PROTEIN, TRAO PROTEIN | | Authors: | Rivera-Calzada, A, Fronzes, R, Savva, C.G, Chandran, V, Lian, P.W, Laeremans, T, Pardon, E, Steyaert, J, Remaut, H, Waksman, G, Orlova, E.V. | | Deposit date: | 2012-11-10 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structure of a Bacterial Type Iv Secretion Core Complex at Subnanometre Resolution.
Embo J., 32, 2013
|
|
3ZBJ
 
 | | Fitting results in the I-layer of the subnanometer structure of the bacterial pKM101 type IV secretion system core complex digested with elastase | | Descriptor: | TRAO PROTEIN | | Authors: | Rivera-Calzada, A, Fronzes, R, Savva, C.G, Chandran, V, Lian, P.W, Laeremans, T, Pardon, E, Steyaert, J, Remaut, H, Waksman, G, Orlova, E.V. | | Deposit date: | 2012-11-10 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structure of a Bacterial Type Iv Secretion Core Complex at Subnanometre Resolution.
Embo J., 32, 2013
|
|
4AQ1
 
 | |
4AQ0
 
 | | Structure of the Gh92 Family Glycosyl Hydrolase Ccman5 in complex with deoxymannojirimycin | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Baranova, E, Tiels, P, Callewaert, N, Remaut, H. | | Deposit date: | 2012-04-12 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A Bacterial Glycosidase Enables Mannose-6-Phosphate Modification and Improved Cellular Uptake of Yeast-Produced Recombinant Human Lysosomal Enzymes.
Nat.Biotechnol., 30, 2012
|
|
4AVH
 
 | | Structure of the FimH lectin domain in the trigonal space group, in complex with a thioalkyl alpha-D-mannoside at 2.1 A resolution | | Descriptor: | 3-(propylsulfanyl)propyl alpha-D-mannopyranoside, FIMH, NICKEL (II) ION, ... | | Authors: | Wellens, A, Lahmann, M, Touaibia, M, Vaucher, J, Oscarson, S, Roy, R, Remaut, H, Bouckaert, J. | | Deposit date: | 2012-05-26 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | The Tyrosine Gate as a Potential Entropic Lever in the Receptor-Binding Site of the Bacterial Adhesin Fimh.
Biochemistry, 51, 2012
|
|
4AVI
 
 | | Structure of the FimH lectin domain in the trigonal space group, in complex with a methyl ester octyl alpha-D-mannoside at 2.4 A resolution | | Descriptor: | FIMH, METHYL ESTER OCTYL ALPHA-1O-D-MANNOPYRANNOSIDE, NICKEL (II) ION, ... | | Authors: | Wellens, A, Lahmann, M, Touaibia, M, Vaucher, J, Oscarson, S, Roy, R, Remaut, H, Bouckaert, J. | | Deposit date: | 2012-05-26 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Tyrosine Gate as a Potential Entropic Lever in the Receptor-Binding Site of the Bacterial Adhesin Fimh.
Biochemistry, 51, 2012
|
|
4AUY
 
 | | Structure of the FimH lectin domain in the trigonal space group, in complex with an hydroxyl propynyl phenyl alpha-D-mannoside at 2.1 A resolution | | Descriptor: | 4-(3-hydroxyprop-1-yn-1-yl)phenyl alpha-D-mannopyranoside, FIMH, NICKEL (II) ION, ... | | Authors: | Wellens, A, Lahmann, M, Touaibia, M, Vaucher, J, Oscarson, S, Roy, R, Remaut, H, Bouckaert, J. | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | The Tyrosine Gate as a Potential Entropic Lever in the Receptor-Binding Site of the Bacterial Adhesin Fimh.
Biochemistry, 51, 2012
|
|
4AV0
 
 | | Structure of the FimH lectin domain in the trigonal space group, in complex with a methoxy phenyl propynyl alpha-D-mannoside at 2.1 A resolution | | Descriptor: | 3-(4-methoxyphenyl)prop-2-yn-1-yl alpha-D-mannopyranoside, FIMH, NICKEL (II) ION, ... | | Authors: | Wellens, A, Lahmann, M, Touaibia, M, Vaucher, J, Oscarson, S, Roy, R, Remaut, H, Bouckaert, J. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | The Tyrosine Gate as a Potential Entropic Lever in the Receptor-Binding Site of the Bacterial Adhesin Fimh.
Biochemistry, 51, 2012
|
|
4AVK
 
 | | Structure of trigonal FimH lectin domain crystal soaked with an alpha- D-mannoside O-linked to propynyl pyridine at 2.4A resolution | | Descriptor: | 3-(pyridin-3-yl)prop-2-yn-1-yl alpha-D-mannopyranoside, FIMH, NICKEL (II) ION, ... | | Authors: | Wellens, A, Lahmann, M, Touaibia, M, Vaucher, J, Oscarson, S, Roy, R, Remaut, H, Bouckaert, J. | | Deposit date: | 2012-05-26 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Tyrosine Gate as a Potential Entropic Lever in the Receptor-Binding Site of the Bacterial Adhesin Fimh.
Biochemistry, 51, 2012
|
|
4AV5
 
 | | Structure of a triclinic crystal of the FimH lectin domain in complex with a propynyl biphenyl alpha-D-mannoside, at 1.4 A resolution | | Descriptor: | (2R,3S,4S,5S,6S)-2-(hydroxymethyl)-6-[3-(4-phenylphenyl)prop-2-ynoxy]oxane-3,4,5-triol, FIMH, SULFATE ION | | Authors: | Wellens, A, Lahmann, M, Touaibia, M, Vaucher, J, Oscarson, S, Roy, R, Remaut, H, Bouckaert, J. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Tyrosine Gate as a Potential Entropic Lever in the Receptor-Binding Site of the Bacterial Adhesin Fimh.
Biochemistry, 51, 2012
|
|
4AVJ
 
 | | Structure of the FimH lectin domain in the trigonal space group, in complex with a methanol triazol ethyl phenyl alpha-D-mannoside at 2.1 A resolution | | Descriptor: | FIMH, METHANOL TRIAZOL ETHYL PHENYL 1O-ALPHA-D-MANNOPYRANOSIDE, NICKEL (II) ION, ... | | Authors: | Wellens, A, Lahmann, M, Touaibia, M, Vaucher, J, Oscarson, S, Roy, R, Remaut, H, Bouckaert, J. | | Deposit date: | 2012-05-26 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | The Tyrosine Gate as a Potential Entropic Lever in the Receptor-Binding Site of the Bacterial Adhesin Fimh.
Biochemistry, 51, 2012
|
|
4AUU
 
 | | Crystal structure of apo FimH lectin domain at 1.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, FIMH, NICKEL (II) ION, ... | | Authors: | Wellens, A, Lahmann, M, Touaibia, M, Vaucher, J, Oscarson, S, Roy, R, Remaut, H, Bouckaert, J. | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Tyrosine Gate as a Potential Entropic Lever in the Receptor-Binding Site of the Bacterial Adhesin Fimh.
Biochemistry, 51, 2012
|
|
5ICJ
 
 | | Crystal structure of the Mycobacterium tuberculosis transcriptional repressor EthR2 in complex with BDM41420 | | Descriptor: | 4,4,4-trifluoro-1-(3-phenyl-1-oxa-2,8-diazaspiro[4.5]dec-2-en-8-yl)butan-1-one, Probable transcriptional regulatory protein | | Authors: | Wohlkonig, A, Remaut, H, Tanina, A, Meyer, F, Willand, N, Baulard, A.R, Wintjens, R. | | Deposit date: | 2016-02-23 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversion of antibiotic resistance in Mycobacterium tuberculosis by spiroisoxazoline SMARt-420.
Science, 355, 2017
|
|
7ZQT
 
 | |
1EAR
 
 | | Crystal structure of Bacillus pasteurii UreE at 1.7 A. Type II crystal form. | | Descriptor: | UREASE ACCESSORY PROTEIN UREE, ZINC ION | | Authors: | Remaut, H, Safarov, N, Ciurli, S, Van Beeumen, J. | | Deposit date: | 2001-07-16 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Ni2+ Transport and Assembly of the Urease Active Site by the Metallochaperone Uree from Bacillus Pasteurii
J.Biol.Chem., 276, 2001
|
|
1EB0
 
 | | Crystal structure of Bacillus pasteurii UreE at 1.85 A, phased by SIRAS. Type I crystal form. | | Descriptor: | UREASE ACCESSORY PROTEIN UREE, ZINC ION | | Authors: | Remaut, H, Safarov, N, Ciurli, S, Van Beeumen, J. | | Deposit date: | 2001-07-17 | | Release date: | 2002-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Ni2+ Transport and Assembly of the Urease Active Site by the Metallochaperone Uree from Bacillus Pasteurii
J.Biol.Chem., 276, 2001
|
|
6XVI
 
 | |
6XUX
 
 | | Crystal structure of Megabody Mb-Nb207-cYgjK_NO | | Descriptor: | CALCIUM ION, Nanobody,Glucosidase YgjK,Glucosidase YgjK,Nanobody | | Authors: | Steyaert, J, Uchanski, T, Fischer, B. | | Deposit date: | 2020-01-21 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.90000641 Å) | | Cite: | Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM.
Nat.Methods, 18, 2021
|
|
6XV8
 
 | |
3OHN
 
 | | Crystal structure of the FimD translocation domain | | Descriptor: | Outer membrane usher protein FimD | | Authors: | Wang, T, Li, H. | | Deposit date: | 2010-08-17 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate.
Nature, 474, 2011
|
|
5VQ5
 
 | | Crystal Structure of the Lectin Domain From the F17-like Adhesin, UclD | | Descriptor: | Adhesin, IODIDE ION | | Authors: | Klein, R.D, Spaulding, C.N, Dodson, K.W, Pinkner, J.S, Hultgren, S.J, Fremont, D. | | Deposit date: | 2017-05-08 | | Release date: | 2017-05-24 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective depletion of uropathogenic E. coli from the gut by a FimH antagonist.
Nature, 546, 2017
|
|
4BWO
 
 | | The FedF adhesin from entrrotoxigenic Escherichia coli is a sulfate- binding lectin | | Descriptor: | BROMIDE ION, F18 FIMBRIAL ADHESIN AC, SULFATE ION | | Authors: | Lonardi, E, Moonens, K, Buts, L, de Boer, A.R, Olsson, J.D.M, Weiss, M.S, Fabre, E, Guerardel, Y, Deelder, A.M, Oscarson, S, Wuhrer, M, Bouckaert, J. | | Deposit date: | 2013-07-03 | | Release date: | 2013-08-28 | | Last modified: | 2014-05-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia Coli
Biology, 2, 2013
|
|
