1K2Y
 
 | |
1K35
 
 | |
1PCJ
 
 | |
1PCM
 
 | |
1P5D
 
 | |
1P5G
 
 | |
2H5A
 
 | | Complex of the enzyme PMM/PGM with xylose 1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-xylopyranose, Phosphomannomutase/phosphoglucomutase, ZINC ION | | Authors: | Regni, C, Shackelford, G.S, Beamer, L.J. | | Deposit date: | 2006-05-25 | | Release date: | 2006-08-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Complexes of the enzyme phosphomannomutase/phosphoglucomutase with a slow substrate and an inhibitor.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2FKF
 
 | |
2FKM
 
 | | PMM/PGM S108D mutant with alpha-d-glucose 1,6-bisphosphate bound | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, Phosphomannomutase/phosphoglucomutase, ZINC ION | | Authors: | Regni, C.A, Beamer, L.J. | | Deposit date: | 2006-01-04 | | Release date: | 2006-04-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The reaction of phosphohexomutase from Pseudomonas aeruginosa: structural insights into a simple processive enzyme.
J.Biol.Chem., 281, 2006
|
|
2H4L
 
 | | Complex of PMM/PGM with ribose 1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-ribofuranose, Phosphomannomutase/phosphoglucomutase, ZINC ION | | Authors: | Beamer, L.J. | | Deposit date: | 2006-05-24 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Complexes of the enzyme phosphomannomutase/phosphoglucomutase with a slow substrate and an inhibitor.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
7M7W
 
 | | Antibodies to the SARS-CoV-2 receptor-binding domain that maximize breadth and resistance to viral escape | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal antibody S2H97 Fab heavy chain, Monoclonal antibody S2H97 Fab light chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7RNJ
 
 | | S2P6 Fab fragment bound to the SARS-CoV/SARS-CoV-2 spike stem helix peptide | | Descriptor: | Monoclonal antibody S2P6 Fab heavy chain, Monoclonal antibody S2P6 Fab light chain, SULFATE ION, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M, Sauer, M.M, Veesler, D. | | Deposit date: | 2021-07-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Broad betacoronavirus neutralization by a stem helix-specific human antibody.
Science, 373, 2021
|
|
8ERR
 
 | |
8ERQ
 
 | |
7R7N
 
 | |
7R6X
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2E12 Fab, S309 Fab, and S304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Monoclonal antibody S2E12 Fab heavy chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7R6W
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2X35 Fab and S309 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Snell, G, Czudnochowski, N, Hernandez, P, Nix, J.C, Croll, T.I, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
8FXB
 
 | | SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
8FXC
 
 | | SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
8S9G
 
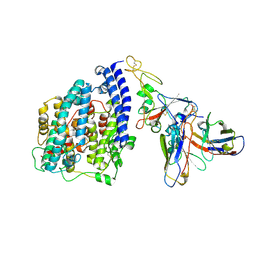 | | SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Fab Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-03-28 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
7N8I
 
 | |
7N8H
 
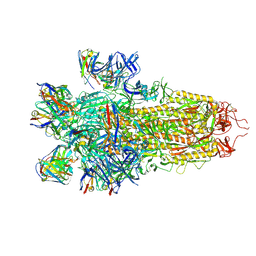 | |
7OH1
 
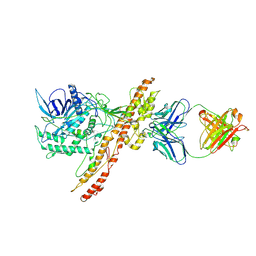 | |
7OH0
 
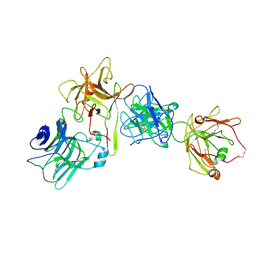 | |
6MJZ
 
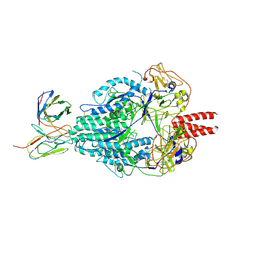 | | Cryo-EM structure of Human Parainfluenza Virus Type 3 (hPIV3) in complex with antibody PIA174 | | Descriptor: | Fusion glycoprotein F0, PIA174 Fab Heavy chain, PIA174 Fab Light chain | | Authors: | Acharya, P, Stewart-Jones, G, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-09-24 | | Release date: | 2018-11-14 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure-based design of a quadrivalent fusion glycoprotein vaccine for human parainfluenza virus types 1-4.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
