2LP7
 
 | |
2M7G
 
 | |
2EXL
 
 | | GRP94 N-terminal Domain bound to geldanamycin | | Descriptor: | Endoplasmin, GELDANAMYCIN, PENTAETHYLENE GLYCOL, ... | | Authors: | Reardon, P.N, Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2005-11-08 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Different poses for ligand and chaperone in inhibitor-bound Hsp90 and GRP94: implications for paralog-specific drug design.
J.Mol.Biol., 388, 2009
|
|
7UI5
 
 | | Evolution avoids a pathological stabilizing interaction in the immune protein S100A9 | | Descriptor: | CALCIUM ION, Protein S100-A9 | | Authors: | Reardon, P.N, Harman, J.L, Costello, S.M, Warren, G.D, Phillips, S.R, Connor, P.J, Marqusee, S, Harms, M.J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Evolution avoids a pathological stabilizing interaction in the immune protein S100A9.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2M7W
 
 | | Independently verified structure of gp41-M-MAT, a membrane associated MPER trimer from HIV-1 gp41 | | Descriptor: | Envelope glycoprotein | | Authors: | Martin, J.W, Reardon, P.N, Sage, H.S, Moses, D.S, Munir, A.S, Haynes, B.F, Spicer, L.D, Donald, B.R. | | Deposit date: | 2013-05-01 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of an HIV-1-neutralizing antibody target, the lipid-bound gp41 envelope membrane proximal region trimer.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2GFD
 
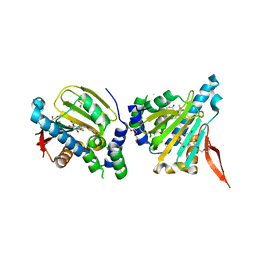 | | GRP94 in complex with the novel HSP90 Inhibitor Radamide | | Descriptor: | Endoplasmin, METHYL 3-CHLORO-2-{3-[(2,5-DIHYDROXY-4-METHOXYPHENYL)AMINO]-3-OXOPROPYL}-4,6-DIHYDROXYBENZOATE, PENTAETHYLENE GLYCOL, ... | | Authors: | Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2006-03-21 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Different poses for ligand and chaperone in inhibitor-bound Hsp90 and GRP94: implications for paralog-specific drug design.
J.Mol.Biol., 388, 2009
|
|
2FXS
 
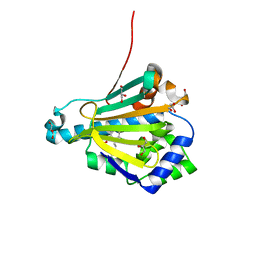 | | Yeast HSP82 in complex with the novel HSP90 Inhibitor Radamide | | Descriptor: | ATP-dependent molecular chaperone HSP82, GLYCEROL, METHYL 3-CHLORO-2-{3-[(2,5-DIHYDROXY-4-METHOXYPHENYL)AMINO]-3-OXOPROPYL}-4,6-DIHYDROXYBENZOATE | | Authors: | Immormino, R.M, Gewirth, D.T. | | Deposit date: | 2006-02-06 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Different poses for ligand and chaperone in inhibitor-bound Hsp90 and GRP94: implications for paralog-specific drug design.
J.Mol.Biol., 388, 2009
|
|
2ESA
 
 | |
3C0E
 
 | |
3C11
 
 | |
