7K35
 
 | | EQADH-NADH-4-METHYLBENZYL ALCOHOL, p21 | | Descriptor: | (4-methylphenyl)methanol, (4R)-2-METHYLPENTANE-2,4-DIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Plapp, B.V, Ramaswamy, S. | | Deposit date: | 2020-09-10 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Alternative binding modes in abortive NADH-alcohol complexes of horse liver alcohol dehydrogenase.
Arch.Biochem.Biophys., 701, 2021
|
|
5VJ5
 
 | | Horse Liver Alcohol Dehydrogenase Complexed with 1,10-Phenanthroline | | Descriptor: | 1,10-PHENANTHROLINE, Alcohol dehydrogenase E chain, ZINC ION | | Authors: | Plapp, B.V, Baskar Raj, S, Ramaswamy, S. | | Deposit date: | 2017-04-18 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Horse Liver Alcohol Dehydrogenase: Zinc Coordination and Catalysis.
Biochemistry, 56, 2017
|
|
1JU9
 
 | |
4JQW
 
 | | Crystal Structure of a Complex of NOD1 CARD and Ubiquitin | | Descriptor: | Nucleotide-binding oligomerization domain-containing protein 1, PHOSPHATE ION, Polyubiquitin-C | | Authors: | Ver Heul, A.M, Gakhar, L, Piper, R.C, Ramaswamy, S. | | Deposit date: | 2013-03-20 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a complex of NOD1 CARD and ubiquitin
Plos One, 9, 2014
|
|
7DA8
 
 | |
1J90
 
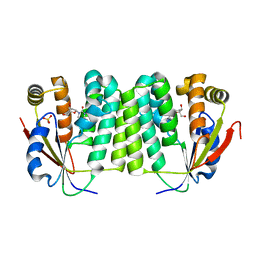 | | Crystal Structure of Drosophila Deoxyribonucleoside Kinase | | Descriptor: | 2'-DEOXYCYTIDINE, Deoxyribonucleoside kinase, SULFATE ION | | Authors: | Johansson, K, Ramaswamy, S, Ljungkrantz, C, Knecht, W, Piskur, J, Munch-Petersen, B, Eriksson, S, Eklund, H. | | Deposit date: | 2001-05-23 | | Release date: | 2001-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis for substrate specificities of cellular deoxyribonucleoside kinases.
Nat.Struct.Biol., 8, 2001
|
|
3F1W
 
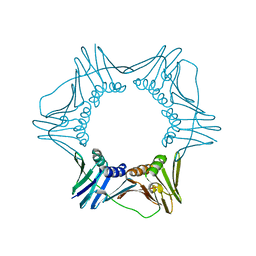 | |
3GPM
 
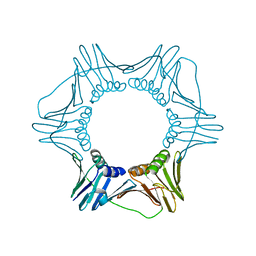 | | Structure of the trimeric form of the E113G PCNA mutant protein | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Freudenthal, B.D, Gakhar, L, Ramaswamy, S, Washington, M.T. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A charged residue at the subunit interface of PCNA promotes trimer formation by destabilizing alternate subunit interactions.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3GPN
 
 | | Structure of the non-trimeric form of the E113G PCNA mutant protein | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Freudenthal, B.D, Gakhar, L, Ramaswamy, S, Washington, M.T. | | Deposit date: | 2009-03-23 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A charged residue at the subunit interface of PCNA promotes trimer formation by destabilizing alternate subunit interactions.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5NVA
 
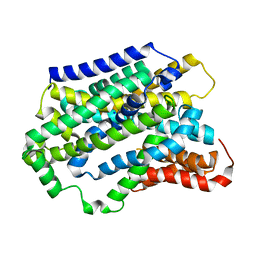 | | Substrate-bound outward-open state of a Na+-coupled sialic acid symporter reveals a novel Na+-site | | Descriptor: | N-acetyl-beta-neuraminic acid, Putative sodium:solute symporter, SODIUM ION | | Authors: | Wahlgren, W.Y, North, R.A, Dunevall, E, Goyal, P, Grabe, M, Dobson, R, Abramson, J, Ramaswamy, S, Friemann, R. | | Deposit date: | 2017-05-03 | | Release date: | 2018-04-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Substrate-bound outward-open structure of a Na+-coupled sialic acid symporter reveals a new Na+site.
Nat Commun, 9, 2018
|
|
5NV9
 
 | | Substrate-bound outward-open state of a Na+-coupled sialic acid symporter reveals a novel Na+-site | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, N-acetyl-beta-neuraminic acid, PHOSPHATE ION, ... | | Authors: | Wahlgren, W.Y, North, R.A, Dunevall, E, Paz, A, Goyal, P, Bisignano, P, Grabe, M, Dobson, R, Abramson, J, Ramaswamy, S, Friemann, R. | | Deposit date: | 2017-05-03 | | Release date: | 2018-04-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate-bound outward-open structure of a Na+-coupled sialic acid symporter reveals a new Na+site.
Nat Commun, 9, 2018
|
|
1A4S
 
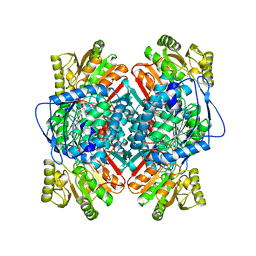 | | BETAINE ALDEHYDE DEHYDROGENASE FROM COD LIVER | | Descriptor: | BETAINE ALDEHYDE DEHYDROGENASE | | Authors: | Johansson, K, El Ahmad, M, Hjelmqvist, L, Ramaswamy, S, Jornvall, H, Eklund, H. | | Deposit date: | 1998-02-03 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of betaine aldehyde dehydrogenase at 2.1 A resolution.
Protein Sci., 7, 1998
|
|
1BPW
 
 | | BETAINE ALDEHYDE DEHYDROGENASE FROM COD LIVER | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (ALDEHYDE DEHYDROGENASE) | | Authors: | Johansson, K, El Ahmad, M, Ramaswamy, S, Hjelmqvist, L, Jornvall, H, Eklund, H. | | Deposit date: | 1998-08-12 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of betaine aldehyde dehydrogenase at 2.1 A resolution.
Protein Sci., 7, 1998
|
|
5F1E
 
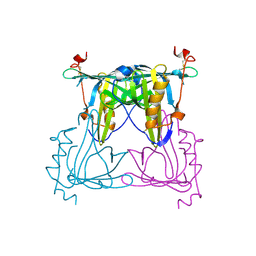 | | Apo protein of Sandercyanin | | Descriptor: | Sandercyanin Fluorescent Protein | | Authors: | Ghosh, S, Ramaswamy, S. | | Deposit date: | 2015-11-30 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Blue protein with red fluorescence
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EZ2
 
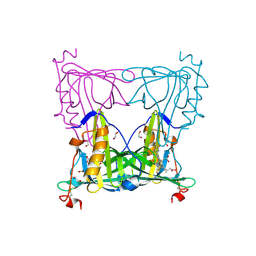 | | Sandercyanin Fluorescent Protein (SFP) | | Descriptor: | 1,2-ETHANEDIOL, BILIVERDINE IX ALPHA, Sandercyanin Fluorescent Protein | | Authors: | Ghosh, S, Ramaswamy, S. | | Deposit date: | 2015-11-26 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Blue protein with red fluorescence
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1EG9
 
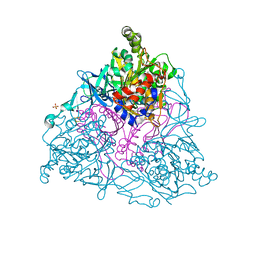 | | NAPHTHALENE 1,2-DIOXYGENASE WITH INDOLE BOUND IN THE ACTIVE SITE. | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, INDOLE, ... | | Authors: | Carredano, E, Karlsson, A, Kauppi, B, Choudhury, D, Parales, R.E, Parales, J.V, Lee, K, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2000-02-15 | | Release date: | 2000-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate binding site of naphthalene 1,2-dioxygenase: functional implications of indole binding.
J.Mol.Biol., 296, 2000
|
|
1QV6
 
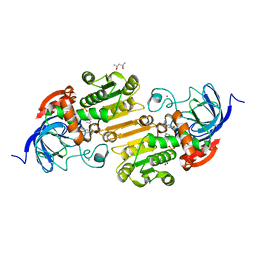 | | HORSE LIVER ALCOHOL DEHYDROGENASE HIS51GLN/LYS228ARG MUTANT COMPLEXED WITH NAD+ AND 2,4-DIFLUOROBENZYL ALCOHOL | | Descriptor: | (2,4-DIFLUOROPHENYL)METHANOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Alcohol dehydrogenase E chain, ... | | Authors: | Lebrun, L.A, Park, D.-H, Ramaswamy, S, Plapp, B.V. | | Deposit date: | 2003-08-26 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Participation of histidine-51 in catalysis by horse liver alcohol dehydrogenase.
Biochemistry, 43, 2004
|
|
1QV7
 
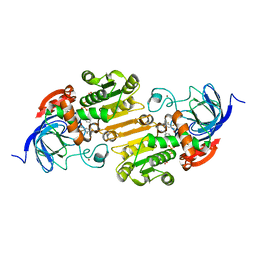 | | HORSE LIVER ALCOHOL DEHYDROGENASE HIS51GLN/LYS228ARG MUTANT COMPLEXED WITH NAD+ AND 2,3-DIFLUOROBENZYL ALCOHOL | | Descriptor: | 2,3-DIFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lebrun, L.A, Park, D.-H, Ramaswamy, S, Plapp, B.V. | | Deposit date: | 2003-08-27 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Participation of histidine-51 in catalysis by horse liver alcohol dehydrogenase.
Biochemistry, 43, 2004
|
|
2QPZ
 
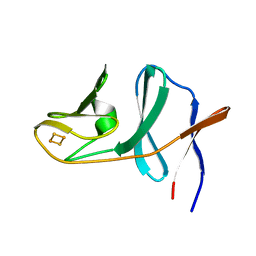 | | Naphthalene 1,2-dioxygenase Rieske ferredoxin | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Naphthalene 1,2-dioxygenase system ferredoxin subunit | | Authors: | Brown, E.N, Ramaswamy, S, Karlsson, A, Friemann, R, Parales, J.V, Parales, R, Gibson, D.T. | | Deposit date: | 2007-07-25 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Determining Rieske cluster reduction potentials.
J.Biol.Inorg.Chem., 13, 2008
|
|
1XSM
 
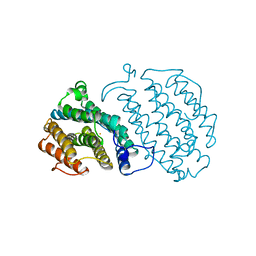 | | PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE FROM MOUSE | | Descriptor: | FE (III) ION, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Kauppi, B, Nielsen, B.N, Ramaswamy, S, Kjoller-Larsen, I, Thelander, M, Thelander, L, Eklund, H. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of mammalian ribonucleotide reductase protein R2 reveals a more-accessible iron-radical site than Escherichia coli R2.
J.Mol.Biol., 262, 1996
|
|
5VKR
 
 | |
2B24
 
 | | Crystal structure of naphthalene 1,2-dioxygenase from Rhodococcus sp. bound to indole | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, INDOLE, ... | | Authors: | Gakhar, L, Malik, Z.A, Allen, C.C, Lipscomb, D.A, Larkin, M.J, Ramaswamy, S. | | Deposit date: | 2005-09-16 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and Increased Thermostability of Rhodococcus sp. Naphthalene 1,2-Dioxygenase.
J.Bacteriol., 187, 2005
|
|
2B1X
 
 | | Crystal structure of naphthalene 1,2-dioxygenase from Rhodococcus sp. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Gakhar, L, Malik, Z.A, Allen, C.C, Lipscomb, D.A, Larkin, M.J, Ramaswamy, S. | | Deposit date: | 2005-09-16 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Increased Thermostability of Rhodococcus sp. Naphthalene 1,2-Dioxygenase.
J.Bacteriol., 187, 2005
|
|
2BMO
 
 | | The Crystal Structure of Nitrobenzene Dioxygenase | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, FE (III) ION, ... | | Authors: | Friemann, R, Ivkovic-Jensen, M.M, Lessner, D.J, Yu, C, Gibson, D.T, Parales, R.E, Eklund, H, Ramaswamy, S. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural insight into the dioxygenation of nitroarene compounds: the crystal structure of nitrobenzene dioxygenase.
J. Mol. Biol., 348, 2005
|
|
2BMQ
 
 | | The Crystal Structure of Nitrobenzene Dioxygenase in complex with nitrobenzene | | Descriptor: | 1,2-ETHANEDIOL, ETHANOL, FE (III) ION, ... | | Authors: | Friemann, R, Ivkovic-Jensen, M.M, Lessner, D.J, Yu, C, Gibson, D.T, Parales, R.E, Eklund, H, Ramaswamy, S. | | Deposit date: | 2005-03-15 | | Release date: | 2005-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into the dioxygenation of nitroarene compounds: the crystal structure of nitrobenzene dioxygenase.
J. Mol. Biol., 348, 2005
|
|
