2PXT
 
 | | Variant 15 of Ribonucleoprotein Core of the E. Coli Signal Recognition Particle | | Descriptor: | 4.5 S RNA, COBALT HEXAMMINE(III), Signal recognition particle protein | | Authors: | Keel, A.Y, Rambo, R.P, Batey, R.T, Kieft, J.S. | | Deposit date: | 2007-05-14 | | Release date: | 2007-08-07 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A General Strategy to Solve the Phase Problem in RNA Crystallography.
Structure, 15, 2007
|
|
6WRF
 
 | | ClpX-ClpP complex bound to GFP-ssrA, recognition complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Fei, X, Sauer, R.T. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis of ClpXP recognition and unfolding of ssrA-tagged substrates.
Elife, 9, 2020
|
|
4LVY
 
 | | Structure of the THF riboswitch bound to pemetrexed | | Descriptor: | 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, THF riboswitch | | Authors: | Trausch, J.J, Batey, R.T. | | Deposit date: | 2013-07-26 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Disconnect between High-Affinity Binding and Efficient Regulation by Antifolates and Purines in the Tetrahydrofolate Riboswitch.
Chem.Biol., 21, 2014
|
|
6WSG
 
 | | ClpX-ClpP complex bound to ssrA-tagged GFP, intermediate complex | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpX, ATP-dependent Clp protease proteolytic subunit, Green fluorescent protein, ... | | Authors: | Fei, X, Sauer, R.T. | | Deposit date: | 2020-04-30 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis of ClpXP recognition and unfolding of ssrA-tagged substrates.
Elife, 9, 2020
|
|
4LVV
 
 | | Structure of the THF riboswitch | | Descriptor: | N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, THF riboswitch | | Authors: | Trausch, J.J, Batey, R.T. | | Deposit date: | 2013-07-26 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Disconnect between High-Affinity Binding and Efficient Regulation by Antifolates and Purines in the Tetrahydrofolate Riboswitch.
Chem.Biol., 21, 2014
|
|
4LW0
 
 | | Structure of the THF riboswitch bound to adenine | | Descriptor: | ADENINE, THF riboswitch | | Authors: | Trausch, J.J, Batey, R.T. | | Deposit date: | 2013-07-26 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | A Disconnect between High-Affinity Binding and Efficient Regulation by Antifolates and Purines in the Tetrahydrofolate Riboswitch.
Chem.Biol., 21, 2014
|
|
6WR2
 
 | |
6PXK
 
 | | 3.65 Angstroms resolution structure of HslU with an axial-channel plug | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, SULFATE ION, ... | | Authors: | Baytshtok, V, Grant, R.A, Sauer, R.T. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.647 Å) | | Cite: | Heat activates the AAA+ HslUV protease by melting an axial autoinhibitory plug.
Cell Rep, 34, 2021
|
|
6PXL
 
 | | 3.74 Angstroms resolution structure of HlsU with an axial-channel plug | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, MAGNESIUM ION, ... | | Authors: | Baytshtok, V, Grant, R.A, Sauer, R.T. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.741 Å) | | Cite: | Heat activates the AAA+ HslUV protease by melting an axial autoinhibitory plug.
Cell Rep, 34, 2021
|
|
6QPX
 
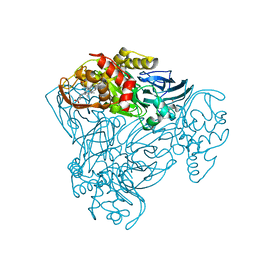 | | Crystal structure of nitrite bound Y323A mutant of haem-Cu containing nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, HEME C, ... | | Authors: | Antonyuk, S.V, Shenoy, R.T, Hedison, T.M, Eady, R.R, Hasnain, S.S, Scrutton, N.S. | | Deposit date: | 2019-02-15 | | Release date: | 2019-11-06 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unexpected Roles of a Tether Harboring a Tyrosine Gatekeeper Residue in Modular Nitrite Reductase Catalysis.
Acs Catalysis, 9, 2019
|
|
6QQ2
 
 | | Crystal structure of nitrite bound Y323F mutant of haem-Cu containing nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, HEME C, ... | | Authors: | Antonyuk, S.V, Shenoy, R.T, Hedison, T.M, Eady, R.R, Hasnain, S.S, Scrutton, N.S. | | Deposit date: | 2019-02-16 | | Release date: | 2019-11-06 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unexpected Roles of a Tether Harboring a Tyrosine Gatekeeper Residue in Modular Nitrite Reductase Catalysis.
Acs Catalysis, 9, 2019
|
|
6QQ1
 
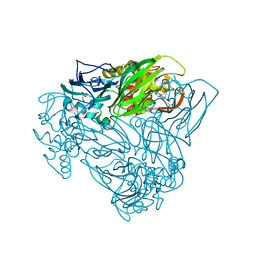 | | Crystal structure of as isolated Y323F mutant of haem-Cu containing nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Shenoy, R.T, Hedison, T.M, Eady, R.R, Hasnain, S.S, Scrutton, N.S. | | Deposit date: | 2019-02-16 | | Release date: | 2019-11-06 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unexpected Roles of a Tether Harboring a Tyrosine Gatekeeper Residue in Modular Nitrite Reductase Catalysis.
Acs Catalysis, 9, 2019
|
|
6R4N
 
 | | Crystal structure of S. cerevisia Niemann-Pick type C protein NPC2 with ergosterol bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ERGOSTEROL, ... | | Authors: | Winkler, M.B.L, Kidmose, R.T, Pedersen, B.P. | | Deposit date: | 2019-03-22 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insight into Eukaryotic Sterol Transport through Niemann-Pick Type C Proteins.
Cell, 179, 2019
|
|
6R4M
 
 | |
6R4L
 
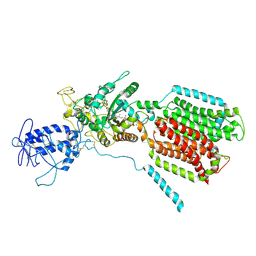 | | Crystal structure of S. cerevisia Niemann-Pick type C protein NCR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ERGOSTEROL, NPC intracellular cholesterol transporter 1-related protein 1, ... | | Authors: | Winkler, M.B.L, Kidmose, R.T, Pedersen, B.P. | | Deposit date: | 2019-03-22 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Insight into Eukaryotic Sterol Transport through Niemann-Pick Type C Proteins.
Cell, 179, 2019
|
|
6QPZ
 
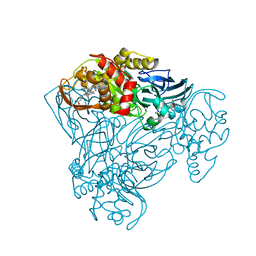 | | Crystal structure of as isolated Y323E mutant of haem-Cu containing nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Shenoy, R.T, Hedison, T.M, Eady, R.R, Hasnain, S.S, Scrutton, N.S. | | Deposit date: | 2019-02-16 | | Release date: | 2019-11-06 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Unexpected Roles of a Tether Harboring a Tyrosine Gatekeeper Residue in Modular Nitrite Reductase Catalysis.
Acs Catalysis, 9, 2019
|
|
6QPV
 
 | | Crystal structure of as isolated Y323A mutant of haem-Cu containing nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, HEME C | | Authors: | Antonyuk, S.V, Shenoy, R.T, Hedison, T.M, Eady, R.R, Hasnain, S.S, Scrutton, N.S. | | Deposit date: | 2019-02-15 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected Roles of a Tether Harboring a Tyrosine Gatekeeper Residue in Modular Nitrite Reductase Catalysis.
Acs Catalysis, 9, 2019
|
|
6QQ0
 
 | | Crystal structure of nitrite bound Y323E mutant of haem-Cu containing nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Shenoy, R.T, Hedison, T.M, Eady, R.R, Hasnain, S.S, Scrutton, N.S. | | Deposit date: | 2019-02-16 | | Release date: | 2019-11-06 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Unexpected Roles of a Tether Harboring a Tyrosine Gatekeeper Residue in Modular Nitrite Reductase Catalysis.
Acs Catalysis, 9, 2019
|
|
966C
 
 | | CRYSTAL STRUCTURE OF FIBROBLAST COLLAGENASE-1 COMPLEXED TO A DIPHENYL-ETHER SULPHONE BASED HYDROXAMIC ACID | | Descriptor: | CALCIUM ION, MMP-1, N-HYDROXY-2-[4-(4-PHENOXY-BENZENESULFONYL)-TETRAHYDRO-PYRAN-4-YL]-ACETAMIDE, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-07 | | Release date: | 1999-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
8U9O
 
 | |
3L0E
 
 | | X-ray crystal structure of a Potent Liver X Receptor Modulator | | Descriptor: | N-(2-chloro-6-fluorobenzyl)-1-methyl-N-{[3'-(methylsulfonyl)biphenyl-4-yl]methyl}-1H-imidazole-4-sulfonamide, Nuclear receptor coactivator 2, Oxysterols receptor LXR-beta | | Authors: | Gampe Jr, R.T. | | Deposit date: | 2009-12-09 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of tertiary sulfonamides as potent liver X receptor antagonists.
J.Med.Chem., 53, 2010
|
|
5KPY
 
 | | Structure of a 5-hydroxytryptophan aptamer | | Descriptor: | 5-hydroxy-L-tryptophan, 5-hydroxytryptophan RNA aptamer, IRIDIUM HEXAMMINE ION, ... | | Authors: | Batey, R.T, Porter, E, Merck, M. | | Deposit date: | 2016-07-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recurrent RNA motifs as scaffolds for genetically encodable small-molecule biosensors.
Nat. Chem. Biol., 13, 2017
|
|
8DOV
 
 | | Crystal structure of the Shr Hemoglobin Interacting Domain 2 (HID2) in complex with Hemoglobin | | Descriptor: | GLYCEROL, Heme-binding protein Shr, Hemoglobin subunit alpha, ... | | Authors: | Macdonald, R, Mahoney, B.J, Cascio, D, Clubb, R.T. | | Deposit date: | 2022-07-14 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Shr receptor from Streptococcus pyogenes uses a cap and release mechanism to acquire heme-iron from human hemoglobin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EET
 
 | |
8EEP
 
 | |
