2J8P
 
 | | NMR structure of C-terminal domain of human CstF-64 | | Descriptor: | CLEAVAGE STIMULATION FACTOR 64 KDA SUBUNIT | | Authors: | Qu, X, Perez-Canadillas, J.M, Agrawal, S, De Baecke, J, Cheng, H, Varani, G, Moore, C. | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-Terminal Domains of Vertebrate Cstf-64 and its Yeast Orthologue RNA15 Form a New Structure Critical for Mrna 3'-End Processing.
J.Biol.Chem., 282, 2007
|
|
7WU5
 
 | | Cryo-EM structure of the adhesion GPCR ADGRF1(H565A/T567A) in complex with miniGi | | Descriptor: | Adhesion G-protein coupled receptor F1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU2
 
 | | Cryo-EM structure of the adhesion GPCR ADGRD1 in complex with miniGs | | Descriptor: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU4
 
 | | Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGi | | Descriptor: | Adhesion G-protein coupled receptor F1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU3
 
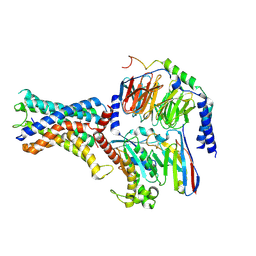 | | Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGs | | Descriptor: | Adhesion G-protein coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-05 | | Release date: | 2022-04-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
8K2S
 
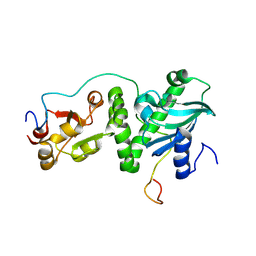 | |
8K2R
 
 | |
8K2T
 
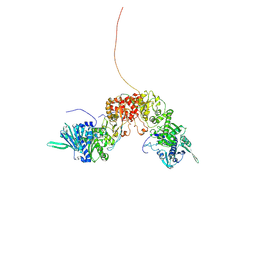 | |
8STB
 
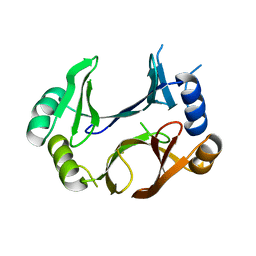 | | The structure of abxF, an enzyme catalyzing the formation of the chiral spiroketal of an anthrabenzoxocinone antibiotic, (-)-ABX | | Descriptor: | GLYCEROL, Glyoxalase, SULFATE ION, ... | | Authors: | Luo, Z, Jia, X, Yan, X, Qu, X, Kobe, B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-05-22 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | An enzymatic dual-oxa Diels-Alder reaction constructs the oxygen-bridged tricyclic acetal unit of (-)-anthrabenzoxocinone.
Nat.Chem., 2025
|
|
8EPY
 
 | | The solution structure of abxF in complex with its product (-)-ABX, an enzyme catalyzing the formation of the chiral spiroketal of an anthrabenzoxocinone antibiotic, (-)-ABX | | Descriptor: | (6R,16R)-3,11,13,15-tetrahydroxy-1,6,9,9-tetramethyl-6,7,9,16-tetrahydro-14H-6,16-epoxyanthra[2,3-e]benzo[b]oxocin-14-one, Glyoxalase | | Authors: | Jia, X, Yan, X, Qu, X, Mobli, M. | | Deposit date: | 2022-10-06 | | Release date: | 2024-04-10 | | Last modified: | 2025-04-23 | | Method: | SOLUTION NMR | | Cite: | An enzymatic dual-oxa Diels–Alder reaction constructs the oxygen-bridged tricyclic acetal unit of (–)-anthrabenzoxocinone
Nat.Chem., 2025
|
|
8EO9
 
 | | The solution structure of abxF, an enzyme catalyzing the formation of chiral spiroketal of an antibiotics, (-)-ABX | | Descriptor: | Glyoxalase | | Authors: | Jia, X, Yan, X, Mobli, M, Qu, X. | | Deposit date: | 2022-10-02 | | Release date: | 2024-04-03 | | Last modified: | 2025-04-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of abxF, an enzyme catalyzing the formation of chiral spiroketal of an antibiotics, (-)-ABX.
Nat.Chem., 2025
|
|
6VZB
 
 | | Crystal structure of cytochrome P450 NasF5053 S284A-V288A mutant variant from Streptomyces sp. NRRL F-5053 in the cyclo-L-Trp-L-Pro-bound state | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6VXV
 
 | | Crystal structure of cyclo-L-Trp-L-Pro-bound cytochrome P450 NasF5053 from Streptomyces sp. NRRL F-5053 | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6W0S
 
 | | Crystal structure of substrate free cytochrome P450 NasF5053 from Streptomyces sp. NRRL F-5053 | | Descriptor: | BROMIDE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-03-02 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
6VZA
 
 | | Crystal structure of cytochrome P450 NasF5053 Q65I-A86G mutant variant from Streptomyces sp. NRRL F-5053 in the cyclo-L-Trp-L-Pro-bound state | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
2LEY
 
 | | Solution structure of (R7G)-Crp4 | | Descriptor: | Alpha-defensin 4 | | Authors: | Rosengren, K, Andersson, H.S, Haugaard-Kedstrom, L.M, Bengtsson, E, Daly, N.L, Figueredo, S.M, Qu, X, Craik, D.J, Ouellette, A.J. | | Deposit date: | 2011-06-26 | | Release date: | 2012-05-16 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The alpha-defensin salt-bridge induces backbone stability to facilitate folding and confer proteolytic resistance.
Amino Acids, 43, 2012
|
|
7XXJ
 
 | | Echo 18 incubated with FcRn at pH5.5 | | Descriptor: | PALMITIC ACID, VP1, VP2, ... | | Authors: | Liu, C.C, Qu, X. | | Deposit date: | 2022-05-30 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Human FcRn Is a Two-in-One Attachment-Uncoating Receptor for Echovirus 18.
Mbio, 13, 2022
|
|
7XXA
 
 | | Complex of Echo 18 and FcRn at pH7.4 | | Descriptor: | IgG receptor FcRn large subunit p51, VP1, VP2, ... | | Authors: | Liu, C.C, Qu, X. | | Deposit date: | 2022-05-29 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Human FcRn Is a Two-in-One Attachment-Uncoating Receptor for Echovirus 18.
Mbio, 13, 2022
|
|
7XXG
 
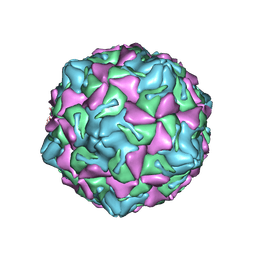 | | Echo 18 at pH5.5 | | Descriptor: | PALMITIC ACID, VP1, VP2, ... | | Authors: | Liu, C.C, Qu, X. | | Deposit date: | 2022-05-30 | | Release date: | 2022-06-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Human FcRn Is a Two-in-One Attachment-Uncoating Receptor for Echovirus 18.
Mbio, 13, 2022
|
|
8GXQ
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in MH-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X, Xu, Y. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.04 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
8GXS
 
 | | PIC-Mediator in complex with +1 nucleosome (T40N) in H-binding state | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Wang, X, Liu, W, Ren, Y, Qu, X, Li, J, Yin, X. | | Deposit date: | 2022-09-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structures of +1 nucleosome-bound PIC-Mediator complex.
Science, 378, 2022
|
|
8K56
 
 | |
8K51
 
 | |
8WAN
 
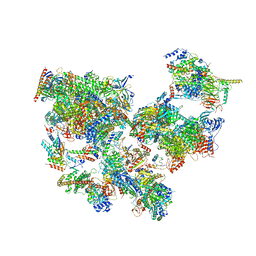 | | Structure of transcribing complex 4 (TC4), the initially transcribing complex with Pol II positioned 4nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.07 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAS
 
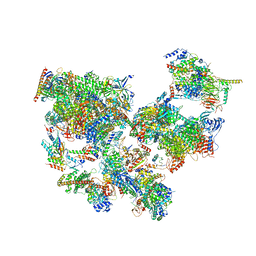 | | Structure of transcribing complex 9 (TC9), the initially transcribing complex with Pol II positioned 9nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.13 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
