2JRC
 
 | |
2MA7
 
 | | Solution NMR Structure of Zinc finger protein Eos from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR7992A | | Descriptor: | ZINC ION, Zinc finger protein Eos | | Authors: | Pulavarti, S.V, Mills, J.L, Wang, D, Jigabm, E, Hamilton, K, Xiao, R, Everett, J.K, Acton, T.B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-28 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Zinc finger protein Eos from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR7992A
To be Published
|
|
2L7K
 
 | | Solution NMR Structure of protein CD1104.2 from Clostridium difficile, Northeast Structural Genomics Consortium Target CfR130 | | Descriptor: | Uncharacterized protein | | Authors: | Pulavarti, S.V.S.R.K, Eletsky, A, Mills, J.L, Sukumaran, D.K, Wang, H, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Lee, H, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-13 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of CfR130 from Clostridium difficile, Northeast Structural Genomics Consortium Target CfR130
To be Published
|
|
2LE1
 
 | | Solution NMR Structure of Tfu_2981 from Thermobifida fusca, Northeast Structural Genomics Consortium Target TfR85A | | Descriptor: | Uncharacterized protein | | Authors: | Pulavarti, S.V.S.R.K, Eletsky, A, Mills, J.L, Sukumaran, D.K, Wang, D, Ciccosanti, C, Hamilton, K, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Lee, H, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-03 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Tfu_2981 from Thermobifida fusca, Northeast Structural Genomics Consortium Target TfR85A
To be Published
|
|
2MRA
 
 | | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR459 | | Descriptor: | De novo designed protein OR459 | | Authors: | Pulavarti, S.V.S.R.K, Kipnis, Y, Sukumaran, D, Maglaqui, M, Janjua, H, Mao, L, Xiao, R, Kornhaber, G, Baker, D, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-07-02 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed protein, Northeast Structural Genomics Consortium (NESG) Target OR459
To be Published
|
|
2ND3
 
 | | Solution structure of the de novo mini protein gEEH_04 | | Descriptor: | De novo mini protein EEH_04 | | Authors: | Pulavarti, S.V, Bahl, C.D, Gilmore, J.M, Eletsky, A, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
2ND2
 
 | | Solution structure of the de novo mini protein gHHH_06 | | Descriptor: | De novo mini protein HHH_06 | | Authors: | Pulavarti, S.V, Eletsky, A, Bahl, C.D, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
5W9F
 
 | | Solution structure of the de novo mini protein gHEEE_02 | | Descriptor: | De novo mini protein gHEEE_02 | | Authors: | Pulavarti, S.V.S.R.K, Shaw, E.A, Bahl, C.D, Garry, B.W, Baker, D, Szyperski, T. | | Deposit date: | 2017-06-23 | | Release date: | 2018-07-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cytosolic expression, solution structures, and molecular dynamics simulation of genetically encodable disulfide-rich de novo designed peptides.
Protein Sci., 27, 2018
|
|
2KVK
 
 | | Solution structure of ADF/cofilin (LDCOF) from Leishmania donovani | | Descriptor: | Actin severing and dynamics regulatory protein | | Authors: | Pathak, P.P, Pulavarti, S.V, Jain, A, Sahasrabuddhe, A.A, Gupta, C.M, Arora, A. | | Deposit date: | 2010-03-16 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ADF/cofilin from Leishmania donovani
J.Struct.Biol., 172, 2010
|
|
5JI4
 
 | | Solution structure of the de novo mini protein gEEHE_02 | | Descriptor: | W37 | | Authors: | Buchko, G.W, Bahl, C.D, Pulavarti, S.V, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
5JHI
 
 | | Solution structure of the de novo mini protein gEHE_06 | | Descriptor: | W35 | | Authors: | Buchko, G.W, Bahl, C.D, Gilmore, J.M, Pulavarti, S.V, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-09-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
2KGY
 
 | | Solution structure of Rv0603 protein from Mycobacterium tuberculosis H37Rv | | Descriptor: | POSSIBLE EXPORTED PROTEIN | | Authors: | Tripathi, S, Pulavarti, S.V.S.R.K, Pathak, P.P, Meher, A.K, Jain, A, Arora, A. | | Deposit date: | 2009-03-23 | | Release date: | 2010-03-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Rv0603 protein from Mycobacterium tuberculosis H37Rv
To be Published
|
|
2L72
 
 | | Solution structure and dynamics of ADF from Toxoplasma gondii (TgADF) | | Descriptor: | Actin depolymerizing factor, putative | | Authors: | Pathak, P.P, Shukla, V.K, Yadav, R, Jain, A, Srivastava, S, Tripathi, S, Pulavarti, S.V.S.R.K, Arora, A. | | Deposit date: | 2010-12-01 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ADF from Toxoplasma gondii
J.Struct.Biol., 176, 2011
|
|
2MIO
 
 | | Solution NMR Structure of SH3 Domain 1 of Rho GTPase-activating Protein 10 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR9129A | | Descriptor: | Rho GTPase-activating protein 10 | | Authors: | Xu, X, Eletsky, A, Pulavarti, S.V.S.R.K, Wang, H, Janjua, H, Xiao, R, Everett, J.K, Sukumaran, D.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-12-16 | | Release date: | 2014-01-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of SH3 Domain 1 of Rho GTPase-activating Protein 10 from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR9129A
To be Published
|
|
2LY9
 
 | | Solution NMR Structure of Homeobox 2 Domain from Human ZHX1 repressor, Northeast Structural Genomics Consortium (NESG) Target HR7907F | | Descriptor: | Zinc fingers and homeoboxes protein 1 | | Authors: | Xu, X, Eletsky, A, Mills, J.L, Pulavarti, S.V.S.R.K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-09-14 | | Release date: | 2012-11-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Homeobox 2 Domain from Human ZHX1 repressor, Northeast Structural Genomics Consortium (NESG) Target HR7907F
To be Published
|
|
2LZ1
 
 | | Solution NMR Structure of the DNA-Binding Domain of Human NF-E2-Related Factor 2, Northeast Structural Genomics Consortium (NESG) Target HR3520O | | Descriptor: | Nuclear factor erythroid 2-related factor 2 | | Authors: | Eletsky, A, Pulavarti, S.V.S.R.K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-09-22 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the DNA-Binding Domain of Human NF-E2-Related Factor 2, Northeast Structural Genomics Consortium (NESG) Target HR3520O
To be Published
|
|
7T2Y
 
 | | X-ray structure of a designed cold unfolding four helix bundle | | Descriptor: | Designed cold unfolding four helix bundle | | Authors: | Harrison, J.S, Kuhlman, B, Szyperski, T, Premkumar, L, Maguire, J, Pulavarti, S, Yuen, S. | | Deposit date: | 2021-12-06 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | From Protein Design to the Energy Landscape of a Cold Unfolding Protein.
J.Phys.Chem.B, 126, 2022
|
|
7T03
 
 | | NMR structure of a designed cold unfolding four helix bundle | | Descriptor: | Cold unfolding four helix bundle | | Authors: | Pulavarti, S, Szyperski, T, Yuen, S, Maguire, J, Griffin, J, Kuhlman, B. | | Deposit date: | 2021-11-29 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | From Protein Design to the Energy Landscape of a Cold Unfolding Protein.
J.Phys.Chem.B, 126, 2022
|
|
5KWX
 
 | |
4Z86
 
 | |
5IKE
 
 | |
5IMB
 
 | |
5JG9
 
 | |
4ZXP
 
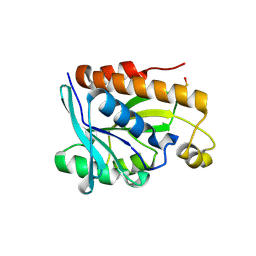 | | Crystal structure of Peptidyl- tRNA Hydrolase from Vibrio cholerae | | Descriptor: | CITRATE ANION, Peptidyl-tRNA hydrolase | | Authors: | Shahid, S, Pal, R.K, Kabra, A, Yadav, R, Kumar, A, Arora, A. | | Deposit date: | 2015-05-20 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Unraveling the stereochemical and dynamic aspects of the catalytic site of bacterial peptidyl-tRNA hydrolase.
RNA, 23, 2017
|
|
5B6J
 
 | |
