6BHS
 
 | | HIV-1 CA hexamer in complex with IP6, hexagonal crystal form | | Descriptor: | Capsid protein p24, INOSITOL HEXAKISPHOSPHATE | | Authors: | Zadrozny, K, Wagner, J.M, Ganser-Pornillos, B.K, Pornillos, O. | | Deposit date: | 2017-10-31 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Inositol phosphates are assembly co-factors for HIV-1.
Nature, 560, 2018
|
|
6BHT
 
 | | HIV-1 CA hexamer in complex with IP6, orthorhombic crystal form | | Descriptor: | Capsid protein p24, INOSITOL HEXAKISPHOSPHATE | | Authors: | Zadrozny, K, Wagner, J.M, Ganser-Pornillos, B.K, Pornillos, O. | | Deposit date: | 2017-10-31 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Inositol phosphates are assembly co-factors for HIV-1.
Nature, 560, 2018
|
|
7R7P
 
 | | Immature HIV-1 CACTD-SP1 lattice with Bevirimat (BVM) and Inositol hexakisphosphate (IP6) | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-22 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
7R7Q
 
 | | Immature HIV-1 CACTD-SP1 lattice with Inositol hexakisphosphate (IP6) | | Descriptor: | Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-22 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
5EIU
 
 | | Mini TRIM5 B-box 2 dimer C2 crystal form | | Descriptor: | TRIM protein-E3 ligase Chimera, ZINC ION | | Authors: | Wagner, J.M, Doss, G, Pornillos, O. | | Deposit date: | 2015-10-30 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | Mechanism of B-box 2 domain-mediated higher-order assembly of the retroviral restriction factor TRIM5 alpha.
Elife, 5, 2016
|
|
5F7T
 
 | | TRIM5 B-box2 and coiled-coil chimera | | Descriptor: | Tripartite motif-containing protein 5,Serine--tRNA ligase,Tripartite motif-containing protein 5, ZINC ION | | Authors: | Wagner, J.M, Doss, G, Pornillos, O. | | Deposit date: | 2015-12-08 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Mechanism of B-box 2 domain-mediated higher-order assembly of the retroviral restriction factor TRIM5 alpha.
Elife, 5, 2016
|
|
7LGC
 
 | |
7LGA
 
 | |
5TSX
 
 | |
5TSV
 
 | |
5TUT
 
 | |
3GV2
 
 | | X-ray Structure of Hexameric HIV-1 CA | | Descriptor: | Capsid protein p24,Carbon dioxide-concentrating mechanism protein CcmK homolog 4 | | Authors: | Kelly, B.N. | | Deposit date: | 2009-03-30 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | X-ray structures of the hexameric building block of the HIV capsid.
Cell(Cambridge,Mass.), 137, 2009
|
|
7ZUD
 
 | | Crystal structure of HIV-1 capsid IP6-CPSF6 complex | | Descriptor: | Capsid protein p24, Cleavage and polyadenylation specificity factor subunit 6, INOSITOL HEXAKISPHOSPHATE | | Authors: | Nicastro, G, Taylor, I.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | CP-MAS and Solution NMR Studies of Allosteric Communication in CA-assemblies of HIV-1.
J.Mol.Biol., 434, 2022
|
|
7TGQ
 
 | |
4WYM
 
 | | Structural basis of HIV-1 capsid recognition by CPSF6 | | Descriptor: | Capsid protein p24, ISOFORM 2 OF CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR SUBUNIT 6 | | Authors: | Battacharya, A, Taylor, A.B, Hart, P.J, Ivanov, D.N. | | Deposit date: | 2014-11-17 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of HIV-1 capsid recognition by PF74 and CPSF6.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7KZH
 
 | |
5VA4
 
 | |
4XFZ
 
 | | Structure of the native full-length HIV-1 capsid protein in complex with PF-3450074 (PF74) | | Descriptor: | CHLORIDE ION, HIV-1 capsid protein, IODIDE ION, ... | | Authors: | Gres, A.T, Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2014-12-29 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | STRUCTURAL VIROLOGY. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability.
Science, 349, 2015
|
|
4XFX
 
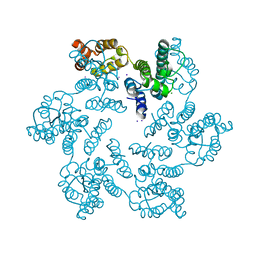 | |
4XFY
 
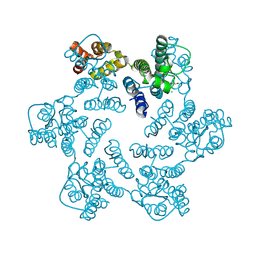 | |
3B61
 
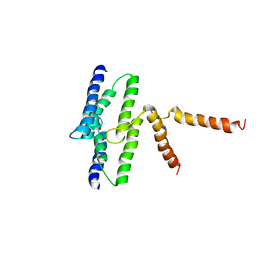 | |
3B5D
 
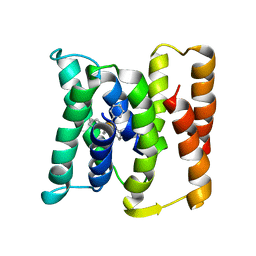 | |
3B62
 
 | |
