3BKC
 
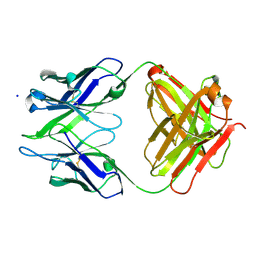 | | Crystal structure of anti-amyloid beta FAB WO2 (P21, FormB) | | Descriptor: | SODIUM ION, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | Deposit date: | 2007-12-06 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
3BKJ
 
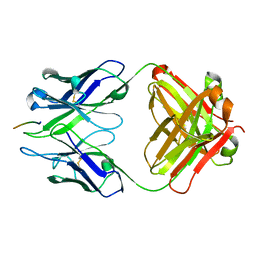 | | Crystal structure of Fab wo2 bound to the n terminal domain of amyloid beta peptide (1-16) | | Descriptor: | Amyloid Beta Peptide, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | Deposit date: | 2007-12-06 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
3BKM
 
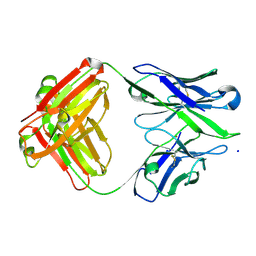 | | Structure of anti-amyloid-beta Fab WO2 (Form A, P212121) | | Descriptor: | SODIUM ION, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa, ... | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | Deposit date: | 2007-12-07 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
1OWT
 
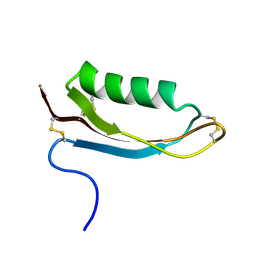 | | Structure of the Alzheimer's disease amyloid precursor protein copper binding domain | | Descriptor: | Amyloid beta A4 protein | | Authors: | Barnham, K.J, McKinstry, W.J, Multhaup, G, Galatis, D, Morton, C.J, Curtain, C.C, Williamson, N.A, White, A.R, Hinds, M.G, Norton, R.S, Beyreuther, K, Masters, C.L, Parker, M.W, Cappai, R. | | Deposit date: | 2003-03-30 | | Release date: | 2003-05-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the Alzheimer's Disease Amyloid Precursor Protein Copper Binding Domain. A REGULATOR OF NEURONAL COPPER HOMEOSTASIS.
J.Biol.Chem., 278, 2003
|
|
1PFO
 
 | | PERFRINGOLYSIN O | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Rossjohn, J, Parker, M.W. | | Deposit date: | 1997-07-31 | | Release date: | 1998-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form.
Cell(Cambridge,Mass.), 89, 1997
|
|
3CWL
 
 | | Crystal structure of alpha-1-antitrypsin, crystal form B | | Descriptor: | Alpha-1-antitrypsin, CHLORIDE ION | | Authors: | Morton, C.J, Hansen, G, Feil, S.C, Adams, J.J, Parker, M.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
1PX6
 
 | | A folding mutant of human class pi glutathione transferase, created by mutating aspartate 153 of the wild-type protein to asparagine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, Glutathione S-transferase P | | Authors: | Kong, G.K.-W, Polekhina, G, McKinstry, W.J, Parker, M.W, Dragani, B, Aceto, A, Paludi, D, Principe, D.R, Mannervik, B, Stenberg, G. | | Deposit date: | 2003-07-02 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The multi-functional role of a highly conserved aspartic acid residue in glutathione transferase P1-1
To be Published
|
|
3CWM
 
 | | Crystal structure of alpha-1-antitrypsin complexed with citrate | | Descriptor: | Alpha-1-antitrypsin, CITRIC ACID | | Authors: | Morton, C.J, Hansen, G, Feil, S.C, Adams, J.J, Parker, M.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
1PX7
 
 | | A folding mutant of human class pi glutathione transferase, created by mutating aspartate 153 of the wild-type protein to glutamate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Kong, G.K.-W, Polekhina, G, McKinstry, W.J, Parker, M.W, Dragani, B, Aceto, A, Paludi, D, Principe, D.R, Mannervik, B, Stenberg, G. | | Deposit date: | 2003-07-02 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The multi-functional role of a highly conserved aspartic acid residue in glutathione transferase P1-1
To be Published
|
|
1PMT
 
 | | GLUTATHIONE TRANSFERASE FROM PROTEUS MIRABILIS | | Descriptor: | GLUTATHIONE, GLUTATHIONE TRANSFERASE | | Authors: | Rossjohn, J, Polekhina, G, Feil, S.C, Allocati, N, Masulli, M, Diilio, C, Parker, M.W. | | Deposit date: | 1998-03-23 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A mixed disulfide bond in bacterial glutathione transferase: functional and evolutionary implications.
Structure, 6, 1998
|
|
3DAQ
 
 | | Crystal structure of dihydrodipicolinate synthase from methicillin-resistant Staphylococcus aureus | | Descriptor: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL | | Authors: | Dobson, R.C.J, Burgess, B.R, Jameson, G.B, Gerrard, J.A, Parker, M.W, Perugini, M.A. | | Deposit date: | 2008-05-29 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and evolution of a novel dimeric enzyme from a clinically-important bacterial pathogen.
J.Biol.Chem., 2008
|
|
4JZJ
 
 | | Crystal Structure of Receptor-Fab Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Broughton, S.E, Parker, M.W. | | Deposit date: | 2013-04-03 | | Release date: | 2014-04-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Dual mechanism of interleukin-3 receptor blockade by an anti-cancer antibody
Cell Rep, 8, 2014
|
|
4FHA
 
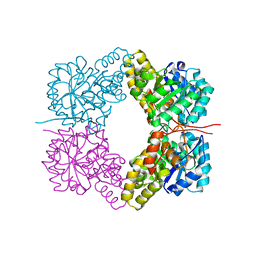 | | Structure of Dihydrodipicolinate Synthase from Streptococcus pneumoniae,bound to pyruvate and lysine | | Descriptor: | Dihydrodipicolinate synthase, LYSINE, SODIUM ION | | Authors: | Perugini, M.A, Dogovski, C, Parker, M.W, Gorman, M.A. | | Deposit date: | 2012-06-06 | | Release date: | 2013-09-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure, Function, Stability and Knockout Phenotype of Dihydrodipicolinate Synthase from Streptococcus pneumoniae
To be Published
|
|
4HIX
 
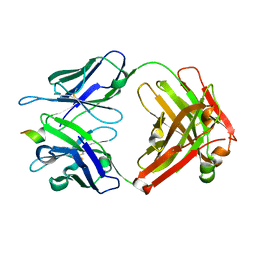 | |
4HSC
 
 | |
4ICN
 
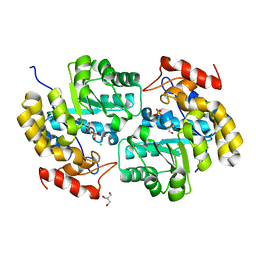 | | Dihydrodipicolinate synthase from shewanella benthica | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DIHYDRODIPICOLINATE SYNTHASE, ... | | Authors: | Wubben, J.M, Paxman, J.J, Dogovski, C, Parker, M.W, Perugini, M.A. | | Deposit date: | 2012-12-10 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cold enzymology offers insight into molecular evolution in quaternary structure
To be Published
|
|
4Y0G
 
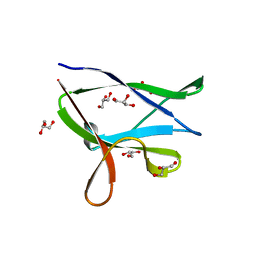 | | beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) | | Descriptor: | 5'-AMP-activated protein kinase subunit beta-2, GLYCEROL | | Authors: | Mobbs, J, Gorman, M.A, Parker, M.W, Gooley, P.R, Griffin, M. | | Deposit date: | 2015-02-06 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Determinants of oligosaccharide specificity of the carbohydrate-binding modules of AMP-activated protein kinase.
Biochem.J., 468, 2015
|
|
4XXD
 
 | | Crystal Structure of mid-region amyloid beta capture by solanezumab | | Descriptor: | Amyloid-beta fragment, Fab Heavy Chain, Fab Light Chain | | Authors: | Hermans, S.J, Crespi, G.A.N, Parker, M.W, Miles, L.A. | | Deposit date: | 2015-01-30 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Molecular basis for mid-region amyloid-beta capture by leading Alzheimer's disease immunotherapies.
Sci Rep, 5, 2015
|
|
4YEF
 
 | | beta1 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclododextrin | | Descriptor: | 5'-AMP-activated protein kinase subunit beta-1, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL, ... | | Authors: | Mobbs, J, Gorman, M.A, Parker, M.W, Gooley, P.R, Griffin, M. | | Deposit date: | 2015-02-24 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Determinants of oligosaccharide specificity of the carbohydrate-binding modules of AMP-activated protein kinase.
Biochem.J., 468, 2015
|
|
4YEE
 
 | | beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin | | Descriptor: | 5'-AMP-activated protein kinase subunit beta-2, Cyclic alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-[alpha-D-glucopyranose-(1-6)]alpha-D-glucopyranose, GLYCEROL | | Authors: | Mobbs, J, Gorman, M.A, Parker, M.W, Gooley, P.R, Griffin, M. | | Deposit date: | 2015-02-24 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of oligosaccharide specificity of the carbohydrate-binding modules of AMP-activated protein kinase.
Biochem.J., 468, 2015
|
|
4ZHX
 
 | | Novel binding site for allosteric activation of AMPK | | Descriptor: | (5S,6R,7R,9R,13cR,14R,16aS)-6-methoxy-5-methyl-7-(methylamino)-6,7,8,9,14,15,16,16a-octahydro-5H,13cH-5,9-epoxy-4b,9a,1 5-triazadibenzo[b,h]cyclonona[1,2,3,4-jkl]cyclopenta[e]-as-indacen-14-ol, 3-[4-(2-hydroxyphenyl)phenyl]-4-oxidanyl-6-oxidanylidene-7H-thieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-activated protein kinase catalytic subunit alpha-2, ... | | Authors: | Langendorf, C.G, Ngoei, K.R, Issa, S.M.A, Ling, N, Gorman, M.A, Parker, M.W, Sakamoto, K, Scott, J.W, Oakhill, J.S, Kemp, B.E. | | Deposit date: | 2015-04-27 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis of allosteric and synergistic activation of AMPK by furan-2-phosphonic derivative C2 binding.
Nat Commun, 7, 2016
|
|
3VQ5
 
 | | HIV-1 IN core domain in complex with N-METHYL-1-(4-METHYL-2-PHENYL-1,3-THIAZOL-5-YL)METHANAMINE | | Descriptor: | CADMIUM ION, CHLORIDE ION, N-methyl-1-(4-methyl-2-phenyl-1,3-thiazol-5-yl)methanamine, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
3VQ4
 
 | | Fragments bound to HIV-1 integrase | | Descriptor: | (5-phenyl-1,2-oxazol-3-yl)methanol, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
3VQ6
 
 | | HIV-1 IN core domain in complex with (1-methyl-5-phenyl-1H-pyrazol-4-yl)methanol | | Descriptor: | (1-methyl-5-phenyl-1H-pyrazol-4-yl)methanol, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
3VQC
 
 | | HIV-1 IN core domain in complex with (5-METHYL-3-PHENYL-1,2-OXAZOL-4-YL)METHANOL | | Descriptor: | (5-methyl-3-phenyl-1,2-oxazol-4-yl)methanol, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
