6PDD
 
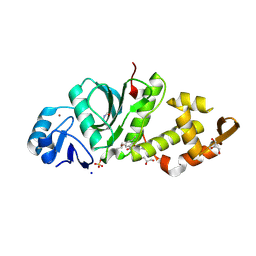 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor 41 | | Descriptor: | 2-fluoro-3-methyl-N'-(phenylsulfonyl)-5-[(propan-2-yl)oxy]benzohydrazide, GLYCEROL, Histone acetyltransferase KAT8, ... | | Authors: | Hermans, S.J, Parker, M.W, Thomas, T, Baell, J.B. | | Deposit date: | 2019-06-18 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of Acylsulfonohydrazide-Derived Inhibitors of the Lysine Acetyltransferase, KAT6A, as Potent Senescence-Inducing Anti-Cancer Agents.
J.Med.Chem., 63, 2020
|
|
3HA4
 
 | |
4Y0G
 
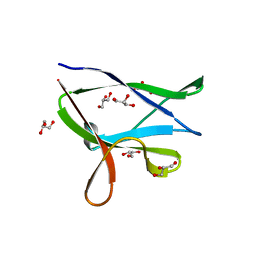 | | beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) | | Descriptor: | 5'-AMP-activated protein kinase subunit beta-2, GLYCEROL | | Authors: | Mobbs, J, Gorman, M.A, Parker, M.W, Gooley, P.R, Griffin, M. | | Deposit date: | 2015-02-06 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Determinants of oligosaccharide specificity of the carbohydrate-binding modules of AMP-activated protein kinase.
Biochem.J., 468, 2015
|
|
4XXD
 
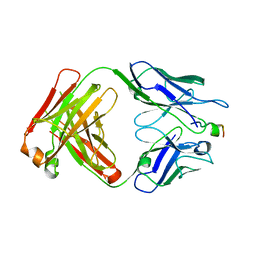 | | Crystal Structure of mid-region amyloid beta capture by solanezumab | | Descriptor: | Amyloid-beta fragment, Fab Heavy Chain, Fab Light Chain | | Authors: | Hermans, S.J, Crespi, G.A.N, Parker, M.W, Miles, L.A. | | Deposit date: | 2015-01-30 | | Release date: | 2015-04-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Molecular basis for mid-region amyloid-beta capture by leading Alzheimer's disease immunotherapies.
Sci Rep, 5, 2015
|
|
4YEF
 
 | | beta1 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclododextrin | | Descriptor: | 5'-AMP-activated protein kinase subunit beta-1, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL, ... | | Authors: | Mobbs, J, Gorman, M.A, Parker, M.W, Gooley, P.R, Griffin, M. | | Deposit date: | 2015-02-24 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Determinants of oligosaccharide specificity of the carbohydrate-binding modules of AMP-activated protein kinase.
Biochem.J., 468, 2015
|
|
4YEE
 
 | | beta2 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclodextrin | | Descriptor: | 5'-AMP-activated protein kinase subunit beta-2, Cyclic alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-[alpha-D-glucopyranose-(1-6)]alpha-D-glucopyranose, GLYCEROL | | Authors: | Mobbs, J, Gorman, M.A, Parker, M.W, Gooley, P.R, Griffin, M. | | Deposit date: | 2015-02-24 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of oligosaccharide specificity of the carbohydrate-binding modules of AMP-activated protein kinase.
Biochem.J., 468, 2015
|
|
3FFD
 
 | | Structure of parathyroid hormone-related protein complexed to a neutralizing monoclonal antibody | | Descriptor: | Monoclonal antibody, heavy chain, Fab fragment, ... | | Authors: | Mckinstry, W.J, Polekhina, G, Diefenbach-Jagger, H, Ho, P.W.M, Sato, K, Onuma, E, Gillespie, M.T, Martin, T.J, Parker, M.W. | | Deposit date: | 2008-12-03 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for antibody discrimination between two hormones that recognize the parathyroid hormone receptor
J.Biol.Chem., 284, 2009
|
|
3CWM
 
 | | Crystal structure of alpha-1-antitrypsin complexed with citrate | | Descriptor: | Alpha-1-antitrypsin, CITRIC ACID | | Authors: | Morton, C.J, Hansen, G, Feil, S.C, Adams, J.J, Parker, M.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
3CWL
 
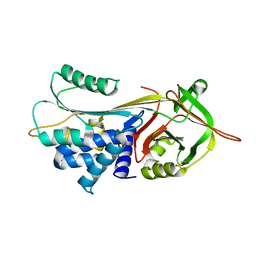 | | Crystal structure of alpha-1-antitrypsin, crystal form B | | Descriptor: | Alpha-1-antitrypsin, CHLORIDE ION | | Authors: | Morton, C.J, Hansen, G, Feil, S.C, Adams, J.J, Parker, M.W. | | Deposit date: | 2008-04-22 | | Release date: | 2008-09-23 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Preventing serpin aggregation: The molecular mechanism of citrate action upon antitrypsin unfolding.
Protein Sci., 17, 2008
|
|
3DAQ
 
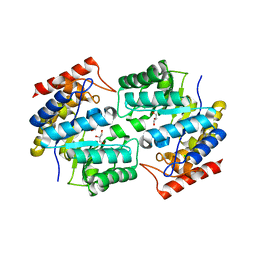 | | Crystal structure of dihydrodipicolinate synthase from methicillin-resistant Staphylococcus aureus | | Descriptor: | CHLORIDE ION, Dihydrodipicolinate synthase, GLYCEROL | | Authors: | Dobson, R.C.J, Burgess, B.R, Jameson, G.B, Gerrard, J.A, Parker, M.W, Perugini, M.A. | | Deposit date: | 2008-05-29 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and evolution of a novel dimeric enzyme from a clinically-important bacterial pathogen.
J.Biol.Chem., 2008
|
|
4ZHX
 
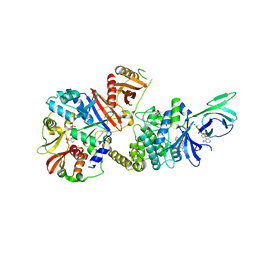 | | Novel binding site for allosteric activation of AMPK | | Descriptor: | (5S,6R,7R,9R,13cR,14R,16aS)-6-methoxy-5-methyl-7-(methylamino)-6,7,8,9,14,15,16,16a-octahydro-5H,13cH-5,9-epoxy-4b,9a,1 5-triazadibenzo[b,h]cyclonona[1,2,3,4-jkl]cyclopenta[e]-as-indacen-14-ol, 3-[4-(2-hydroxyphenyl)phenyl]-4-oxidanyl-6-oxidanylidene-7H-thieno[2,3-b]pyridine-5-carbonitrile, 5'-AMP-activated protein kinase catalytic subunit alpha-2, ... | | Authors: | Langendorf, C.G, Ngoei, K.R, Issa, S.M.A, Ling, N, Gorman, M.A, Parker, M.W, Sakamoto, K, Scott, J.W, Oakhill, J.S, Kemp, B.E. | | Deposit date: | 2015-04-27 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis of allosteric and synergistic activation of AMPK by furan-2-phosphonic derivative C2 binding.
Nat Commun, 7, 2016
|
|
6BA2
 
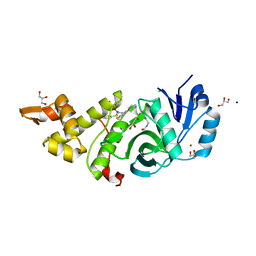 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor | | Descriptor: | 4-fluoro-5-methyl-N'-(phenylsulfonyl)[1,1'-biphenyl]-3-carbohydrazide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hermans, S.J, Chung, M.C, Peat, T.S, Baell, J.B, Thomas, T, Parker, M.W. | | Deposit date: | 2017-10-11 | | Release date: | 2018-08-01 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.85003817 Å) | | Cite: | Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth.
Nature, 560, 2018
|
|
6BFS
 
 | | The mechanism of GM-CSF inhibition by human GM-CSF auto-antibodies | | Descriptor: | Fab Heavy Chain, Fab light Chain, Granulocyte-macrophage colony-stimulating factor | | Authors: | Dhagat, U, Hercus, T.R, Broughton, S.E, Nero, T.L, Lopez, A.F, Parker, M.W. | | Deposit date: | 2017-10-26 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The mechanism of GM-CSF inhibition by human GM-CSF auto-antibodies suggests novel therapeutic opportunities.
MAbs, 10, 2018
|
|
6B2E
 
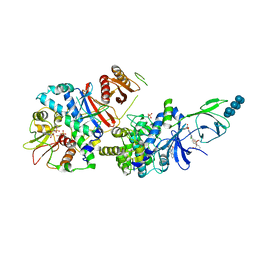 | | Structure of full length human AMPK (a2b2g1) in complex with a small molecule activator SC4. | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Ngoei, K.R.W, Langendorf, C.G, Ling, N.X.Y, Hoque, A, Johnson, S, Camerino, M.C, Walker, S.R, Bozikis, Y.E, Dite, T.A, Ovens, A.J, Smiles, W.J, Jacobs, R, Huang, H, Parker, M.W, Scott, J.W, Rider, M.H, Kemp, B.E, Foitzik, R.C, Baell, J.B, Oakhill, J.S. | | Deposit date: | 2017-09-19 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Determinants for Small-Molecule Activation of Skeletal Muscle AMPK alpha 2 beta 2 gamma 1 by the Glucose Importagog SC4.
Cell Chem Biol, 25, 2018
|
|
6B1U
 
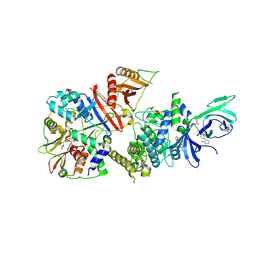 | | Structure of full-length human AMPK (a2b1g1) in complex with a small molecule activator SC4 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Ngoei, K.R.W, Langendorf, C.G, Ling, N.X.Y, Hoque, A, Johnson, S, Camerino, M.C, Walker, S.R, Bozikis, Y.E, Dite, T.A, Ovens, A.J, Smiles, W.J, Jacobs, R, Huang, H, Parker, M.W, Scott, J.W, Rider, M.H, Kemp, B.E, Foitzik, R.C, Baell, J.B, Oakhill, J.S. | | Deposit date: | 2017-09-19 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural Determinants for Small-Molecule Activation of Skeletal Muscle AMPK alpha 2 beta 2 gamma 1 by the Glucose Importagog SC4.
Cell Chem Biol, 25, 2018
|
|
6BA4
 
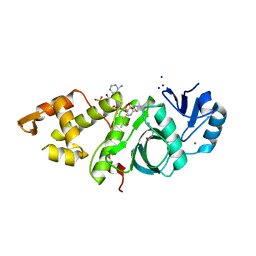 | | Crystal structure of MYST acetyltransferase domain in complex with Acetyl-CoA cofactor | | Descriptor: | ACETYL COENZYME *A, Histone acetyltransferase KAT8, S-{(3S,5R,9R)-1-[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} ethanethioate, ... | | Authors: | Hermans, S.J, Chung, M.C, Peat, T.S, Baell, J.B, Thomas, T, Parker, M.W. | | Deposit date: | 2017-10-12 | | Release date: | 2018-08-01 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth.
Nature, 560, 2018
|
|
1MD3
 
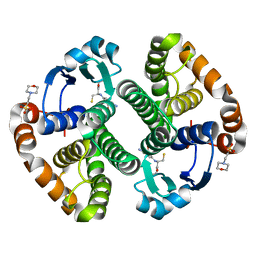 | | A folding mutant of human class pi glutathione transferase, created by mutating glycine 146 of the wild-type protein to alanine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, pi glutathione transferase | | Authors: | Kong, G.K.-W, Dragani, B, Aceto, A, Cocco, R, Mannervik, B, Stenberg, G, McKinstry, W.J, Polekhina, G, Parker, M.W. | | Deposit date: | 2002-08-06 | | Release date: | 2002-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Contribution of Glycine 146 to a Conserved Folding Module Affecting Stability and Refolding of Human Glutathione Transferase P1-1
J.Biol.Chem., 278, 2003
|
|
1MWP
 
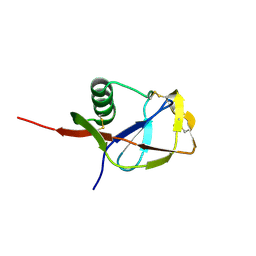 | | N-TERMINAL DOMAIN OF THE AMYLOID PRECURSOR PROTEIN | | Descriptor: | AMYLOID A4 PROTEIN | | Authors: | Rossjohn, J, Cappai, R, Feil, S.C, Henry, A, McKinstry, W.J, Galatis, D, Hesse, L, Multhaup, G, Beyreuther, K, Masters, C.L, Parker, M.W. | | Deposit date: | 1999-03-09 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the N-terminal, growth factor-like domain of Alzheimer amyloid precursor protein.
Nat.Struct.Biol., 6, 1999
|
|
1OWT
 
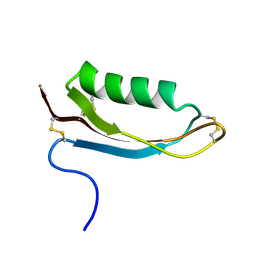 | | Structure of the Alzheimer's disease amyloid precursor protein copper binding domain | | Descriptor: | Amyloid beta A4 protein | | Authors: | Barnham, K.J, McKinstry, W.J, Multhaup, G, Galatis, D, Morton, C.J, Curtain, C.C, Williamson, N.A, White, A.R, Hinds, M.G, Norton, R.S, Beyreuther, K, Masters, C.L, Parker, M.W, Cappai, R. | | Deposit date: | 2003-03-30 | | Release date: | 2003-05-13 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the Alzheimer's Disease Amyloid Precursor Protein Copper Binding Domain. A REGULATOR OF NEURONAL COPPER HOMEOSTASIS.
J.Biol.Chem., 278, 2003
|
|
1PFO
 
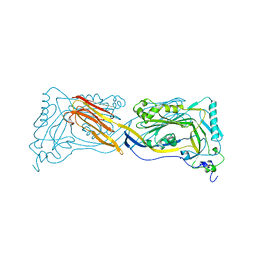 | | PERFRINGOLYSIN O | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Rossjohn, J, Parker, M.W. | | Deposit date: | 1997-07-31 | | Release date: | 1998-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form.
Cell(Cambridge,Mass.), 89, 1997
|
|
1PX6
 
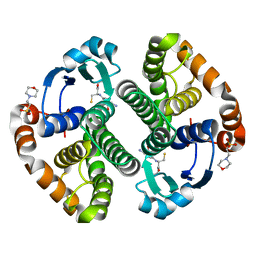 | | A folding mutant of human class pi glutathione transferase, created by mutating aspartate 153 of the wild-type protein to asparagine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, Glutathione S-transferase P | | Authors: | Kong, G.K.-W, Polekhina, G, McKinstry, W.J, Parker, M.W, Dragani, B, Aceto, A, Paludi, D, Principe, D.R, Mannervik, B, Stenberg, G. | | Deposit date: | 2003-07-02 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The multi-functional role of a highly conserved aspartic acid residue in glutathione transferase P1-1
To be Published
|
|
1PX7
 
 | | A folding mutant of human class pi glutathione transferase, created by mutating aspartate 153 of the wild-type protein to glutamate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Kong, G.K.-W, Polekhina, G, McKinstry, W.J, Parker, M.W, Dragani, B, Aceto, A, Paludi, D, Principe, D.R, Mannervik, B, Stenberg, G. | | Deposit date: | 2003-07-02 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The multi-functional role of a highly conserved aspartic acid residue in glutathione transferase P1-1
To be Published
|
|
1PMT
 
 | | GLUTATHIONE TRANSFERASE FROM PROTEUS MIRABILIS | | Descriptor: | GLUTATHIONE, GLUTATHIONE TRANSFERASE | | Authors: | Rossjohn, J, Polekhina, G, Feil, S.C, Allocati, N, Masulli, M, Diilio, C, Parker, M.W. | | Deposit date: | 1998-03-23 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A mixed disulfide bond in bacterial glutathione transferase: functional and evolutionary implications.
Structure, 6, 1998
|
|
4ONI
 
 | |
4PJ6
 
 | | Crystal Structure of Human Insulin Regulated Aminopeptidase with Lysine in Active Site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSINE, ... | | Authors: | Hermans, S.J, Ascher, D.B, Hancock, N.C, Holien, J.K, Michell, B, Morton, C.J, Parker, M.W. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal structure of human insulin-regulated aminopeptidase with specificity for cyclic peptides.
Protein Sci., 24, 2015
|
|
