7QLO
 
 | |
7M78
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | 著者 | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | 登録日 | 2021-03-26 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
7M75
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | 著者 | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | 登録日 | 2021-03-26 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
7M76
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | 著者 | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | 登録日 | 2021-03-26 | | 公開日 | 2021-11-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
7BYO
 
 | |
7BYP
 
 | |
5GV5
 
 | | Crystal structure of Candida antarctica Lipase B with active Ser105 modified with a phosphonate inhibitor | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase B, ... | | 著者 | Park, S.Y, Lee, H. | | 登録日 | 2016-09-02 | | 公開日 | 2017-09-13 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Structural and Experimental Evidence for the Enantiomeric Recognition toward a Bulky sec-Alcohol by Candida antarctica Lipase B
Acs Catalysis, 6, 2016
|
|
6J43
 
 | | Proteinase K determined by PAL-XFEL | | 分子名称: | CALCIUM ION, Proteinase K | | 著者 | Lee, S.J, Park, J. | | 登録日 | 2019-01-07 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Application of a high-throughput microcrystal delivery system to serial femtosecond crystallography.
J.Appl.Crystallogr., 53, 2020
|
|
8SD1
 
 | |
8SD6
 
 | |
8SD7
 
 | |
8SF1
 
 | |
8SD9
 
 | |
8SD8
 
 | |
7D04
 
 | |
7D05
 
 | |
7D01
 
 | |
7D02
 
 | |
6INT
 
 | | xylose isomerase from Paenibacillus sp. R4 | | 分子名称: | CALCIUM ION, Xylose isomerase | | 著者 | Lee, J.H, Lee, C.W, Park, S. | | 登録日 | 2018-10-26 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.942 Å) | | 主引用文献 | Crystal Structure and Functional Characterization of a Xylose Isomerase (PbXI) from the Psychrophilic Soil Microorganism, Paenibacillus sp.
J. Microbiol. Biotechnol., 29, 2019
|
|
7XL9
 
 | | The structure of HucR with urate | | 分子名称: | CHLORIDE ION, Transcriptional regulator, MarR family, ... | | 著者 | Park, S.Y. | | 登録日 | 2022-04-21 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | The structure of Deinococcus radiodurans transcriptional regulator HucR retold with the urate bound.
Biochem.Biophys.Res.Commun., 615, 2022
|
|
8GUY
 
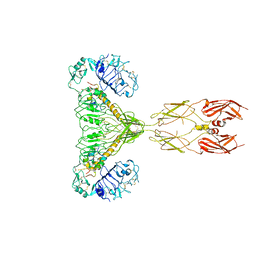 | | human insulin receptor bound with two insulin molecules | | 分子名称: | Insulin A chain, Insulin, isoform 2, ... | | 著者 | Kim, J, Yunn, N, Ryu, S, Cho, Y. | | 登録日 | 2022-09-14 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.18 Å) | | 主引用文献 | Functional selectivity of insulin receptor revealed by aptamer-trapped receptor structures.
Nat Commun, 13, 2022
|
|
6W1S
 
 | | Atomic model of the mammalian Mediator complex | | 分子名称: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | 著者 | Zhao, H, Young, N, Asturias, F. | | 登録日 | 2020-03-04 | | 公開日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (4.02 Å) | | 主引用文献 | A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers.
Cell, 178, 2019
|
|
4GKF
 
 | | Crystal structure and characterization of Cmr5 protein from Pyrococcus furiosus | | 分子名称: | CRISPR system Cmr subunit Cmr5 | | 著者 | Park, J, Sun, J, Park, S, Hwang, H, Park, M, Shin, M.S. | | 登録日 | 2012-08-11 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of Cmr5 from Pyrococcus furiosus and its functional implications
Febs Lett., 587, 2013
|
|
6JCG
 
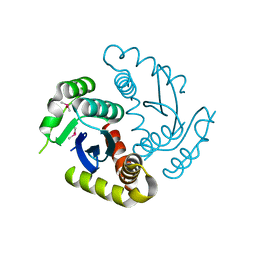 | | Room temperature structure of HIV-1 Integrase catalytic core domain by serial femtosecond crystallography. | | 分子名称: | CACODYLATE ION, Integrase | | 著者 | Park, J.H, Shi, Y, Han, J, Li, X, Kim, T.H, Yun, J.H. | | 登録日 | 2019-01-28 | | 公開日 | 2019-07-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
6JCF
 
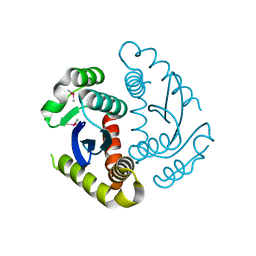 | | Cryogenic structure of HIV-1 Integrase catalytic core domain by synchrotron | | 分子名称: | CACODYLATE ION, Integrase | | 著者 | Park, J.H, Han, J, Kim, T.H, Yun, J.H, Lee, W. | | 登録日 | 2019-01-28 | | 公開日 | 2019-07-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.153 Å) | | 主引用文献 | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
