1MVQ
 
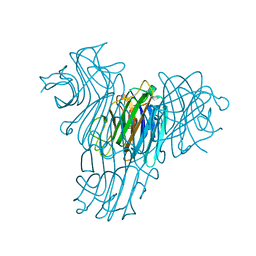 | | Cratylia mollis lectin (isoform 1) in complex with methyl-alpha-D-mannose | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, lectin, ... | | Authors: | de Souza, G.A, Oliveira, P.S, Trapani, S, Correia, M.T, Oliva, G, Coelho, L.C, Greene, L.J. | | Deposit date: | 2002-09-26 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Amino acid sequence and tertiary structure of Cratylia mollis seed lectin
Glycobiology, 13, 2003
|
|
7KO8
 
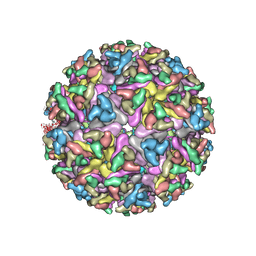 | | Cryo-EM structure of the mature and infective Mayaro virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein, E1 glycoprotein, ... | | Authors: | Riberio-Filho, H.V, Coimbra, L.D, Cassago, A, Rocha, R.P.F, Padilha, A.C.M, Schatz, M, van Heel, M.G, Portugal, R.V, Trivella, D.B.B, de Oliveira, P.S.L, Marques, R.E. | | Deposit date: | 2020-11-06 | | Release date: | 2021-04-28 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the mature and infective Mayaro virus at 4.4 angstrom resolution reveals features of arthritogenic alphaviruses.
Nat Commun, 12, 2021
|
|
6N4C
 
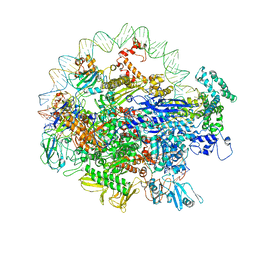 | | EM structure of the DNA wrapping in bacterial open transcription initiation complex | | Descriptor: | DNA (94-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Florez-Ariza, A, Cassago, A, de Oliveira, P.S.L, Guerra, D.G, Portugal, R.V. | | Deposit date: | 2018-11-19 | | Release date: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Interactions of Upstream and Downstream Promoter Regions with RNA Polymerase are Energetically Coupled and a Target of Regulation in Transcription Initiation
Biorxiv, 2020
|
|
4WN5
 
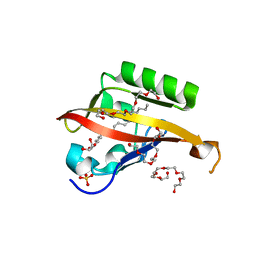 | | Crystal structure of the C-terminal Per-Arnt-Sim (PASb) of human HIF-3alpha9 bound to 18:1-1-monoacylglycerol | | Descriptor: | HEXAETHYLENE GLYCOL, Hypoxia-inducible factor 3-alpha, MONOVACCENIN, ... | | Authors: | Fala, A.M, Oliveira, J.F, Dias, S.M, Ambrosio, A.L. | | Deposit date: | 2014-10-10 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Unsaturated fatty acids as high-affinity ligands of the C-terminal Per-ARNT-Sim domain from the Hypoxia-inducible factor 3 alpha.
Sci Rep, 5, 2015
|
|
7UXX
 
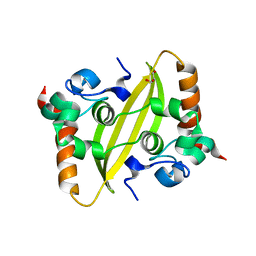 | | Crystal structure of SARS-CoV-2 nucleocapsid protein C-terminal domain | | Descriptor: | ACETATE ION, GLYCEROL, Nucleoprotein | | Authors: | Bezerra, E.H.S, Tonoli, C.C.C, Soprano, A.S, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.
Sci Rep, 12, 2022
|
|
7UXZ
 
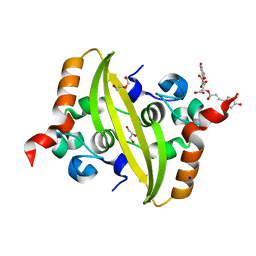 | | Crystal structure of SARS-CoV-2 nucleocapsid protein C-terminal domain complexed with Chicoric acid | | Descriptor: | (2R,3R)-2,3-bis{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}butanedioic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bezerra, E.H.S, Tonoli, C.C.C, Soprano, A.S, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.
Sci Rep, 12, 2022
|
|
4JKT
 
 | | Crystal structure of mouse Glutaminase C, BPTES-bound form | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide) | | Authors: | Fornezari, C, Ferreira, A.P.S, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2013-03-11 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Active Glutaminase C Self-assembles into a Supratetrameric Oligomer That Can Be Disrupted by an Allosteric Inhibitor.
J.Biol.Chem., 288, 2013
|
|
3SUJ
 
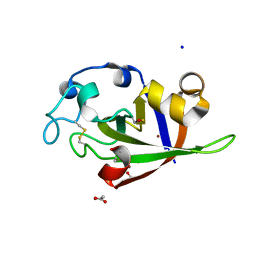 | | Crystal structure of cerato-platanin 1 from M. perniciosa (MpCP1) | | Descriptor: | ACETATE ION, CHLORIDE ION, Cerato-platanin 1, ... | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
3SUK
 
 | | Crystal structure of cerato-platanin 2 from M. perniciosa (MpCP2) | | Descriptor: | Cerato-platanin-like protein | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
3SUM
 
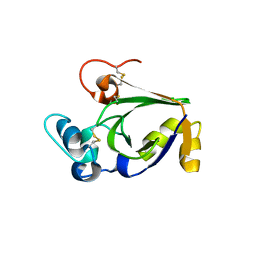 | | Crystal structure of cerato-platanin 5 from M. perniciosa (MpCP5) | | Descriptor: | Cerato-platanin-like protein | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
3SUL
 
 | | Crystal structure of cerato-platanin 3 from M. perniciosa (MpCP3) | | Descriptor: | Cerato-platanin-like protein | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
5DT5
 
 | | Crystal structure of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7 in space group P21 | | Descriptor: | Beta-glucosidase, SULFATE ION | | Authors: | Zanphorlin, L.M, Giuseppe, P.O, Tonoli, C.C.C, Murakami, M.T. | | Deposit date: | 2015-09-17 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Oligomerization as a strategy for cold adaptation: Structure and dynamics of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7.
Sci Rep, 6, 2016
|
|
5DT7
 
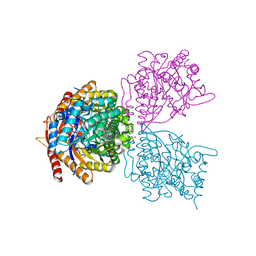 | | Crystal structure of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7 in space group C2221 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Zanphorlin, L.M, Giuseppe, P.O, Tonoli, C.C.C, Murakami, M.T. | | Deposit date: | 2015-09-17 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oligomerization as a strategy for cold adaptation: Structure and dynamics of the GH1 beta-glucosidase from Exiguobacterium antarcticum B7.
Sci Rep, 6, 2016
|
|
6BYC
 
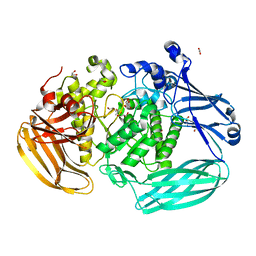 | | Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri | | Descriptor: | ACETATE ION, Beta-mannosidase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Domingues, M.N, Vieira, P.S, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2017-12-20 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Structural basis of exo-beta-mannanase activity in the GH2 family.
J. Biol. Chem., 293, 2018
|
|
6BYE
 
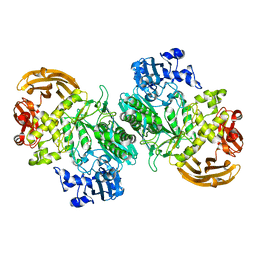 | | Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose | | Descriptor: | ACETATE ION, Beta-mannosidase, beta-D-mannopyranose | | Authors: | Domingues, M.N, Vieira, P.S, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2017-12-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.126 Å) | | Cite: | Structural basis of exo-beta-mannanase activity in the GH2 family.
J. Biol. Chem., 293, 2018
|
|
6BYG
 
 | | Crystal structure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri | | Descriptor: | Beta-mannosidase, beta-D-mannopyranose | | Authors: | Domingues, M.N, Vieira, P.S, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2017-12-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural basis of exo-beta-mannanase activity in the GH2 family.
J. Biol. Chem., 293, 2018
|
|
6BYI
 
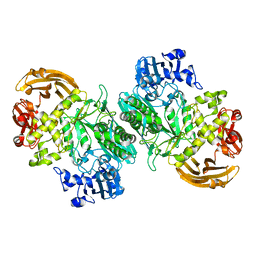 | | Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri | | Descriptor: | Beta-mannosidase, beta-D-mannopyranose | | Authors: | Domingues, M.N, Vieira, P.S, Morais, M.A.B, Murakami, M.T. | | Deposit date: | 2017-12-20 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of exo-beta-mannanase activity in the GH2 family.
J. Biol. Chem., 293, 2018
|
|
4PMX
 
 | | Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri in the native form | | Descriptor: | CALCIUM ION, Xylanase | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4PN2
 
 | | Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylotriose | | Descriptor: | CALCIUM ION, Xylanase, beta-D-xylopyranose | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4PMV
 
 | | Crystal structure of a novel reducing-end xylose-releasing exo-oligoxylanase (XynA) belonging to GH10 family (space group P43212) | | Descriptor: | Endo-1,4-beta-xylanase A | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4PMY
 
 | | Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylose | | Descriptor: | CALCIUM ION, GLYCEROL, Xylanase, ... | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4PMU
 
 | | Crystal structure of a novel reducing-end xylose-releasing exo-oligoxylanase (XynA) belonging to GH10 family (space group P1211) | | Descriptor: | Endo-1,4-beta-xylanase A | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.857 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4PMZ
 
 | | Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylobiose | | Descriptor: | CALCIUM ION, Xylanase, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4J5L
 
 | | Structure of the Cargo Binding Domain from Human Myosin Va | | Descriptor: | SULFATE ION, Unconventional myosin-Va | | Authors: | Nascimento, A.F.Z, Trindade, D.M, Tonoli, C.C.C, Assis, L.H.P, Mahajan, P, Berridge, G, Krojer, T, Vollmar, M, Burgess-Brown, N, von Delft, F, Murakami, M.T. | | Deposit date: | 2013-02-08 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into Functional Overlapping and Differentiation among Myosin V Motors.
J.Biol.Chem., 288, 2013
|
|
4J5M
 
 | | Structure of the Cargo Binding Domain from Human Myosin Vb | | Descriptor: | NITRATE ION, Unconventional myosin-Vb | | Authors: | Nascimento, A.F.Z, Trindade, D.M, Mahajan, P, Berridge, G, Krojer, T, Vollmar, M, Tonoli, C.C.C, Assis, L.H.P, Burgess-Brown, N, von Delft, F, Murakami, M.T. | | Deposit date: | 2013-02-08 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Insights into Functional Overlapping and Differentiation among Myosin V Motors.
J.Biol.Chem., 288, 2013
|
|
