4QYR
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsE KS3 | | Descriptor: | ACETIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Kim, Y, Li, H, Endres, M, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4OQJ
 
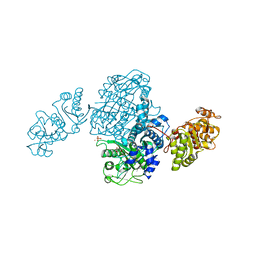 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmQ KS1 | | Descriptor: | GLYCEROL, PHOSPHATE ION, PKS, ... | | Authors: | Nocek, B, Mack, J, Endras, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-09 | | Release date: | 2014-03-19 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4TKT
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS6 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.4289 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5VO3
 
 | |
8D5H
 
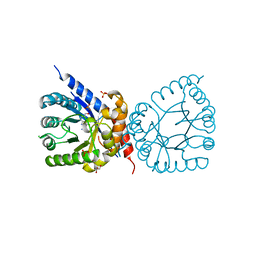 | | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stogios, P.J, Skarina, T, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of dihydropteroate synthase (folP-SMZ_B27) from soil uncultured bacterium in complex with 6-hydroxymethyl-7,8-dihydropterin
To Be Published
|
|
4EYQ
 
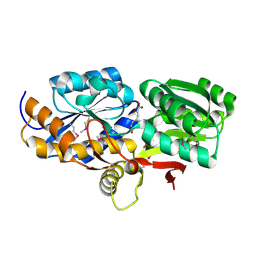 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 in complex with caffeic acid/3-(4-HYDROXY-PHENYL)PYRUVIC ACID | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, CAFFEIC ACID, Extracellular ligand-binding receptor | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4EYO
 
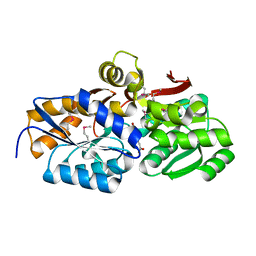 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris HaA2 in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Extracellular ligand-binding receptor | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4I1D
 
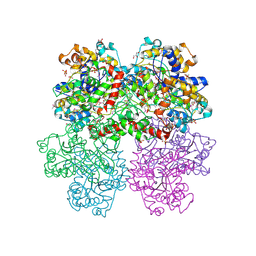 | | The crystal structure of an ABC transporter substrate-binding protein from Bradyrhizobium japonicum USDA 110 | | Descriptor: | ABC transporter substrate-binding protein, ACETATE ION, D-MALATE, ... | | Authors: | Fan, Y, Tan, K, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-05 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4JJX
 
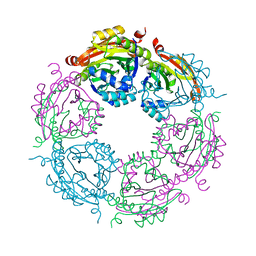 | | Dodecameric structure of spermidine N-acetyltransferase SpeG from Vibrio cholerae O1 biovar eltor | | Descriptor: | Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-08 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
4JLY
 
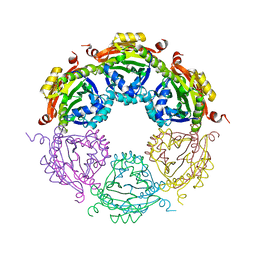 | | Dodecameric structure of spermidine N-acetyltransferase from Vibrio cholerae | | Descriptor: | SULFATE ION, Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
4F8J
 
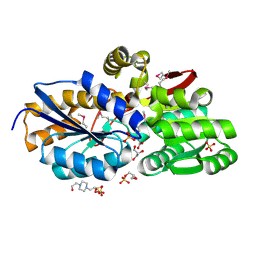 | | The structure of an aromatic compound transport protein from Rhodopseudomonas palustris in complex with p-coumarate | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Cuff, M.E, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-17 | | Release date: | 2012-09-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4FB4
 
 | | The Structure of an ABC-Transporter Family Protein from Rhodopseudomonas palustris in Complex with Caffeic Acid | | Descriptor: | CAFFEIC ACID, GLYCEROL, Putative branched-chain amino acid transport system substrate-binding protein | | Authors: | Cuff, M.E, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-22 | | Release date: | 2012-09-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
1ZKI
 
 | | Structure of conserved protein PA5202 from Pseudomonas aeruginosa | | Descriptor: | ACETIC ACID, hypothetical protein PA5202 | | Authors: | Cuff, M.E, Evdokimova, E, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and activity of the Pseudomonas aeruginosa hotdog-fold thioesterases PA5202 and PA2801.
Biochem.J., 444, 2012
|
|
4MHD
 
 | | Crystal structure of spermidine N-acetyltransferase from Vibrio cholerae in complex with spermidine | | Descriptor: | SPERMIDINE, Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-29 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
4MI4
 
 | | Crystal structure of spermidine N-acetyltransferase from Vibrio cholerae in complex with spermine | | Descriptor: | SPERMINE, Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-30 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
4R87
 
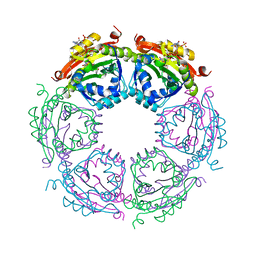 | | Crystal structure of spermidine N-acetyltransferase from Vibrio cholerae in complex with CoA and spermine | | Descriptor: | COENZYME A, DI(HYDROXYETHYL)ETHER, SPERMINE, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-29 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
4R57
 
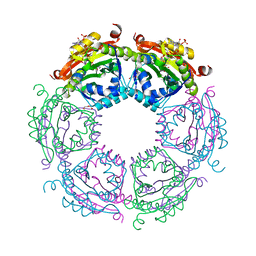 | | Crystal structure of spermidine N-acetyltransferase from Vibrio cholerae in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, DI(HYDROXYETHYL)ETHER, Spermidine n1-acetyltransferase, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | A Novel Polyamine Allosteric Site of SpeG from Vibrio cholerae Is Revealed by Its Dodecameric Structure.
J.Mol.Biol., 427, 2015
|
|
4RXI
 
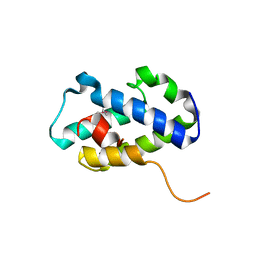 | | Structure of C-terminal domain of uncharacterized protein from Legionella pneumophila | | Descriptor: | hypothetical protein lpg0944 | | Authors: | Cuff, M, Nocek, B, Evdokimova, E, Egorova, O, Joachimiak, A, Ensminger, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-11 | | Release date: | 2015-05-06 | | Last modified: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol Syst Biol, 12, 2016
|
|
4RXV
 
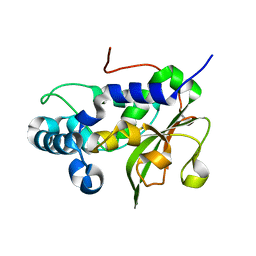 | | The crystal structure of the N-terminal fragment of uncharacterized protein from Legionella pneumophila | | Descriptor: | hypothetical protein lpg0944 | | Authors: | Nocek, B, Cuff, M, Evdokimova, E, Egorova, O, Joachimiak, A, Ensminger, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-12 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol Syst Biol, 12, 2016
|
|
4DQD
 
 | | The crystal structure of a transporter in complex with 3-phenylpyruvic acid | | Descriptor: | 3-HYDROXYPYRUVIC ACID, 3-PHENYLPYRUVIC ACID, Extracellular ligand-binding receptor, ... | | Authors: | Tan, K, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-02-15 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
4HFV
 
 | | Crystal structure of lpg1851 protein from Legionella pneumophila (putative T4SS effector) | | Descriptor: | CITRIC ACID, SUCCINIC ACID, Uncharacterized protein | | Authors: | Michalska, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-10-05 | | Release date: | 2012-11-07 | | Last modified: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol Syst Biol, 12, 2016
|
|
3UKJ
 
 | | Crystal structure of extracellular ligand-binding receptor from Rhodopseudomonas palustris HaA2 | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, Extracellular ligand-binding receptor, GLYCEROL, ... | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-11-09 | | Release date: | 2011-11-23 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
7KZW
 
 | | Crystal structure of FTT_1639c from Francisella tularensis str. tularensis SCHU S4 | | Descriptor: | CHLORIDE ION, FTT_1639c | | Authors: | Stogios, P.J, Skarina, T, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-10 | | Release date: | 2020-12-30 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of FTT_1639c from Francisella tularensis str. tularensis SCHU S4
To Be Published
|
|
7LAO
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-IIb | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aminoglycoside N(3)-acetyltransferase III, MAGNESIUM ION | | Authors: | Stogios, P.J, Evdokimova, E, Osipiuk, J, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-06 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
7UUN
 
 | | Crystal structure of aminoglycoside resistance enzyme ApmA, complex with neomycin | | Descriptor: | 1,2-ETHANEDIOL, Aminocyclitol acetyltransferase ApmA, NEOMYCIN | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Osipiuk, J, Bordeleau, E, Wright, G.D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Mechanistic plasticity in ApmA enables aminoglycoside promiscuity for resistance.
Nat.Chem.Biol., 20, 2024
|
|
