2O7D
 
 | | Tyrosine ammonia-lyase from Rhodobacter sphaeroides, complexed with caffeate | | Descriptor: | CAFFEIC ACID, Putative histidine ammonia-lyase | | Authors: | Louie, G.V, Bowman, M.E, Moffitt, M.C, Baiga, T.J, Moore, B.S, Noel, J.P. | | Deposit date: | 2006-12-10 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases.
Chem.Biol., 13, 2006
|
|
1YQD
 
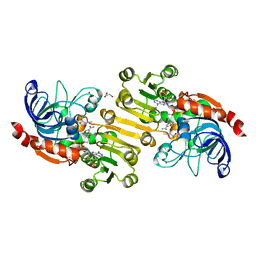 | | Sinapyl Alcohol Dehydrogenase complexed with NADP+ | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Bomati, E.K, Noel, J.P. | | Deposit date: | 2005-02-01 | | Release date: | 2005-07-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Kinetic Basis for Substrate Selectivity in Populus tremuloides Sinapyl Alcohol Dehydrogenase.
Plant Cell, 17, 2005
|
|
1YQX
 
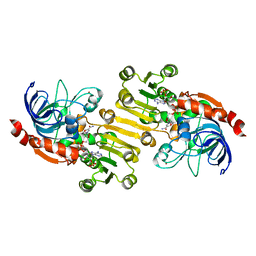 | | Sinapyl Alcohol Dehydrogenase at 2.5 Angstrom Resolution | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION, sinapyl alcohol dehydrogenase | | Authors: | Bomati, E.K, Noel, J.P. | | Deposit date: | 2005-02-02 | | Release date: | 2005-07-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Kinetic Basis for Substrate Selectivity in Populus tremuloides Sinapyl Alcohol Dehydrogenase.
Plant Cell, 17, 2005
|
|
2QZZ
 
 | | Structure of Eugenol Synthase from Ocimum basilicum | | Descriptor: | Eugenol synthase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ethyl (1S,2S)-2-(4-hydroxy-3-methoxyphenyl)cyclopropanecarboxylate | | Authors: | Louie, G.V, Noel, J.P, Bowman, M.E. | | Deposit date: | 2007-08-17 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and reaction mechanism of basil eugenol synthase
Plos One, 2, 2007
|
|
1ZDY
 
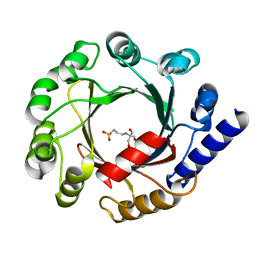 | |
1ZHF
 
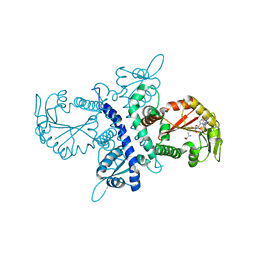 | | Crystal structure of selenomethionine substituted isoflavanone 4'-O-methyltransferase | | Descriptor: | Isoflavanone 4'-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liu, C.-J, Deavours, B.E, Richard, S, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-25 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses.
Plant Cell, 18, 2006
|
|
2O7B
 
 | | Tyrosine ammonia-lyase from Rhodobacter sphaeroides, complexed with coumarate | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Putative histidine ammonia-lyase | | Authors: | Louie, G.V, Bowman, M.E, Moffitt, M.C, Baiga, T.J, Moore, B.S, Noel, J.P. | | Deposit date: | 2006-12-10 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural determinants and modulation of substrate specificity in phenylalanine-tyrosine ammonia-lyases.
Chem.Biol., 13, 2006
|
|
1ZCW
 
 | | Co-crystal structure of Orf2 an aromatic prenyl transferase from Streptomyces sp. strain CL190 complexed with GPP | | Descriptor: | Aromatic prenyltransferase, GERANYL DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kuzuyama, T, Noel, J.P, Richard, S.B. | | Deposit date: | 2005-04-13 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products.
Nature, 435, 2005
|
|
2R2G
 
 | | Structure of Eugenol Synthase from Ocimum basilicum complexed with EMDF | | Descriptor: | Eugenol synthase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ethyl (1S,2S)-2-(4-hydroxy-3-methoxyphenyl)cyclopropanecarboxylate | | Authors: | Louie, G.V, Noel, J.P, Bowman, M.E. | | Deposit date: | 2007-08-24 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and reaction mechanism of basil eugenol synthase
Plos One, 2, 2007
|
|
1ZGJ
 
 | | Crystal structure of isoflavanone 4'-O-methyltransferase complexed with (+)-pisatin | | Descriptor: | (6AR,12AR)-3-(HYDROXYMETHYL)-6H-[1,3]DIOXOLO[5,6][1]BENZOFURO[3,2-C]CHROMEN-6A(12AH)-OL, Isoflavanone 4'-O-methyltransferase', S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liu, C.-J, Deavours, B.E, Richard, S, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-21 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses.
Plant Cell, 18, 2006
|
|
2QYS
 
 | |
1ZGD
 
 | | Chalcone Reductase Complexed With NADP+ at 1.7 Angstrom Resolution | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, chalcone reductase | | Authors: | Bomati, E.K, Austin, M.B, Bowman, M.E, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-21 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural elucidation of chalcone reductase and implications for deoxychalcone biosynthesis
J.Biol.Chem., 280, 2005
|
|
1ZG3
 
 | | Crystal structure of the isoflavanone 4'-O-methyltransferase complexed with SAH and 2,7,4'-trihydroxyisoflavanone | | Descriptor: | (2S,3R)-2,7-DIHYDROXY-3-(4-HYDROXYPHENYL)-2,3-DIHYDRO-4H-CHROMEN-4-ONE, S-ADENOSYL-L-HOMOCYSTEINE, isoflavanone 4'-O-methyltransferase | | Authors: | Liu, C.-J, Deavours, B.E, Richard, S, Ferrer, J.-L, Dixon, R.A, Noel, J.P. | | Deposit date: | 2005-04-20 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for dual functionality of isoflavonoid O-methyltransferases in the evolution of plant defense responses.
Plant Cell, 18, 2006
|
|
1ZDW
 
 | | Co-crystal structure of Orf2 an aromatic prenyl transferase from Streptomyces sp. strain CL190 complexed with GSPP and Flaviolin | | Descriptor: | Aromatic prenyltransferase, FLAVIOLIN, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Kuzuyama, T, Noel, J.P, Richard, S.B. | | Deposit date: | 2005-04-15 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products.
Nature, 435, 2005
|
|
2Q6K
 
 | | SalL with adenosine | | Descriptor: | ADENOSINE, DI(HYDROXYETHYL)ETHER, chlorinase | | Authors: | Pojer, F, Noel, J.P. | | Deposit date: | 2007-06-05 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and characterization of a marine bacterial SAM-dependent chlorinase
Nat.Chem.Biol., 4, 2008
|
|
2Q6L
 
 | | SalL double mutant Y70T/G131S with CLDA and L-MET | | Descriptor: | 5'-CHLORO-5'-DEOXYADENOSINE, Hypothetical protein, METHIONINE | | Authors: | Pojer, F, Noel, J.P. | | Deposit date: | 2007-06-05 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Discovery and characterization of a marine bacterial SAM-dependent chlorinase
Nat.Chem.Biol., 4, 2008
|
|
1ND7
 
 | | Conformational Flexibility Underlies Ubiquitin Ligation Mediated by the WWP1 HECT domain E3 Ligase | | Descriptor: | WW domain-containing protein 1 | | Authors: | Verdecia, M.A, Joaziero, C.A.P, Wells, N.J, Ferrer, J.-L, Bowman, M.E, Hunter, T, Noel, J.P. | | Deposit date: | 2002-12-08 | | Release date: | 2003-09-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Flexibility Underlies Ubiquitin Ligation Mediated by the WWP1 HECT domain E3 Ligase
Mol.Cell, 11, 2003
|
|
1P15
 
 | |
1P13
 
 | | Crystal Structure of the Src SH2 Domain Complexed with Peptide (SDpYANFK) | | Descriptor: | CACODYLATE ION, Peptide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Sonnenburg, E.D, Bilwes, A, Hunter, T, Noel, J.P. | | Deposit date: | 2003-04-11 | | Release date: | 2003-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | The structure of the membrane distal phosphatase domain of RPTPalpha reveals interdomain flexibility and an SH2 domain interaction region.
Biochemistry, 42, 2003
|
|
1OSH
 
 | | A Chemical, Genetic, and Structural Analysis of the nuclear bile acid receptor FXR | | Descriptor: | Bile acid receptor, METHYL 3-{3-[(CYCLOHEXYLCARBONYL){[4'-(DIMETHYLAMINO)BIPHENYL-4-YL]METHYL}AMINO]PHENYL}ACRYLATE | | Authors: | Downes, M, Verdecia, M.A, Roecker, A.J, Hughes, R, Hogenesch, J.B, Kast-Woelbern, H.R, Bowman, M.E, Ferrer, J.-L, Anisfeld, A.M, Edwards, P.A, Rosenfeld, J.M, Alvarez, J.G.A, Noel, J.P, Nicolaou, K.C, Evans, R.M. | | Deposit date: | 2003-03-19 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR
Mol.Cell, 11, 2003
|
|
1M6E
 
 | | CRYSTAL STRUCTURE OF SALICYLIC ACID CARBOXYL METHYLTRANSFERASE (SAMT) | | Descriptor: | 2-HYDROXYBENZOIC ACID, LUTETIUM (III) ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zubieta, C, Ross, J.R, Koscheski, P, Yang, Y, Pichersky, E, Noel, J.P. | | Deposit date: | 2002-07-16 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Substrate Recognition in The Salicylic Acid Carboxyl Methyltransferase Family
Plant Cell, 15, 2003
|
|
1U0U
 
 | | An Aldol Switch Discovered in Stilbene Synthases Mediates Cyclization Specificity of Type III Polyketide Synthases: Pine stilbene synthase structure | | Descriptor: | Dihydropinosylvin synthase | | Authors: | Austin, M.B, Bowman, M.E, Ferrer, J.-L, Schroder, J, Noel, J.P. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | An Aldol Switch Discovered in Stilbene Synthases Mediates Cyclization Specificity of Type III Polyketide Synthases
Chem.Biol., 11, 2004
|
|
1QLV
 
 | |
1U0W
 
 | | An Aldol Switch Discovered in Stilbene Synthases Mediates Cyclization Specificity of Type III Polyketide Synthases: 18xCHS+resveratrol Structure | | Descriptor: | Chalcone synthase 2, RESVERATROL | | Authors: | Austin, M.B, Bowman, M.E, Ferrer, J.-L, Schroder, J, Noel, J.P. | | Deposit date: | 2004-07-14 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Aldol Switch Discovered in Stilbene Synthases Mediates Cyclization Specificity of Type III Polyketide Synthases
Chem.Biol., 11, 2004
|
|
1U0M
 
 | | Crystal Structure of 1,3,6,8-Tetrahydroxynaphthalene Synthase (THNS) from Streptomyces coelicolor A3(2): a Bacterial Type III Polyketide Synthase (PKS) Provides Insights into Enzymatic Control of Reactive Polyketide Intermediates | | Descriptor: | GLYCEROL, POLYETHYLENE GLYCOL (N=34), putative polyketide synthase | | Authors: | Austin, M.B, Izumikawa, M, Bowman, M.E, Udwary, D.W, Ferrer, J.L, Moore, B.S, Noel, J.P. | | Deposit date: | 2004-07-13 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of a bacterial type III polyketide synthase and enzymatic control of reactive polyketide intermediates
J.Biol.Chem., 279, 2004
|
|
