6LEI
 
 | |
6LED
 
 | |
6LE2
 
 | | Structure of D-carbamoylase mutant from Nitratireductor indicus | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, N-carbamoyl-D-amino-acid hydrolase | | 著者 | Ni, Y, Liu, Y.F, Xu, G.C, Dai, W. | | 登録日 | 2019-11-23 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Structure-Guided Engineering of D-Carbamoylase Reveals a Key Loop at Substrate Entrance Tunnel
Acs Catalysis, 10, 2020
|
|
5HSI
 
 | | Crystal structure of tyrosine decarboxylase at 1.73 Angstroms resolution | | 分子名称: | MAGNESIUM ION, Putative decarboxylase | | 著者 | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | 登録日 | 2016-01-25 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.732 Å) | | 主引用文献 | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
5HSJ
 
 | | Structure of tyrosine decarboxylase complex with PLP at 1.9 Angstroms resolution | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Putative decarboxylase | | 著者 | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | 登録日 | 2016-01-25 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
8J43
 
 | | Reductive aminase RA29-WT | | 分子名称: | 1,2-ETHANEDIOL, Beta-hydroxyacid dehydrogenase, 3-hydroxyisobutyrate dehydrogenase, ... | | 著者 | Zheng, X.Y, Xu, G.C, Ni, Y. | | 登録日 | 2023-04-19 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Dynamic kinetic reductive resolution of cyclic keto esters by newly identified stereo complementary reductive aminases.
To Be Published
|
|
8J44
 
 | | Reductive Aminase RA34-WT | | 分子名称: | 6-phosphogluconate dehydrogenase NAD-binding protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zheng, X.Y, Xu, G.C, Ni, Y. | | 登録日 | 2023-04-19 | | 公開日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Dynamic kinetic reductive resolution of cyclic keto esters by newly identified stereo complementary reductive aminases.
To Be Published
|
|
7C3V
 
 | | Structure of a thermostable Alcohol dehydrogenase from Kluyveromyces polyspora(KpADH) | | 分子名称: | Alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Dai, W, Ni, Y, Xu, G, Liu, Y, Wang, Y, Zhou, J. | | 登録日 | 2020-05-14 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.20042944 Å) | | 主引用文献 | Structure of a thermostable Alcohol dehydrogenase from Kluyveromyces polyspora(KpADH)
To Be Published
|
|
6LCG
 
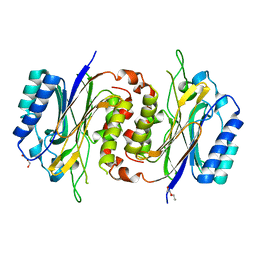 | | Structure of D-carbamoylase mutant from Nitratireductor indicus | | 分子名称: | DI(HYDROXYETHYL)ETHER, N-carbamoyl-D-amino-acid hydrolase | | 著者 | Liu, Y.F, Ni, Y, Xu, G.C, Dai, W. | | 登録日 | 2019-11-18 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure-Guided Engineering of D-Carbamoylase Reveals a Key Loop at Substrate Entrance Tunnel
Acs Catalysis, 10, 2020
|
|
7C1E
 
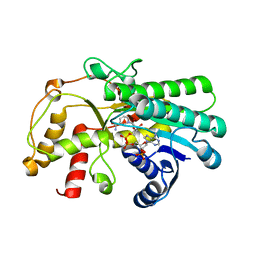 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (Y127W) | | 分子名称: | Epimerase domain-containing protein, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wu, Y.F, Zhou, J.Y, Liu, Y.F, Xu, G.C, Ni, Y. | | 登録日 | 2020-05-03 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Engineering an Alcohol Dehydrogenase from Kluyveromyces polyspora for Efficient Synthesis of Ibrutinib Intermediate
Adv.Synth.Catal., 2021
|
|
8I99
 
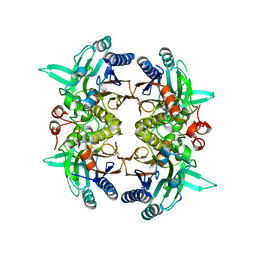 | |
5ZED
 
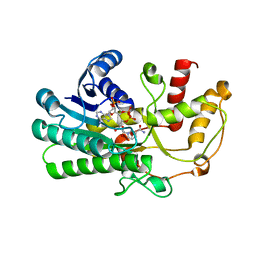 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (E214V/T215S) | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized protein ADH | | 著者 | Wang, Y, Zhou, J.Y, Hou, X.D, Xu, G.C, Wu, L, Rao, Y.J, ZHou, J.H, Ni, Y. | | 登録日 | 2018-02-27 | | 公開日 | 2019-01-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
5Z2X
 
 | | Structure of Alcohol dehydrogenase from Kluyveromyces polyspora(KpADH) | | 分子名称: | 1,2-ETHANEDIOL, Alcohol dehydrogenase, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Wang, Y, Zhou, J.Y, Hou, X.D, Xu, G.C, Wu, L, Rao, Y.J, ZHou, J.H, Ni, Y. | | 登録日 | 2018-01-04 | | 公開日 | 2019-01-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
5ZEC
 
 | | Crystal structure of Kluyveromyces polyspora ADH (KpADH) mutant (Q136N/F161V/S196G/E214G/S237C) | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ETHANOL, ... | | 著者 | Wang, Y, ZHou, J.Y, Hou, X.D, Xu, G.C, Rao, Y.J, Wu, L, Zhou, J.H, Ni, Y. | | 登録日 | 2018-02-27 | | 公開日 | 2019-01-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.779 Å) | | 主引用文献 | Structural Insight into Enantioselective Inversion of an Alcohol Dehydrogenase Reveals a "Polar Gate" in Stereorecognition of Diaryl Ketones.
J. Am. Chem. Soc., 140, 2018
|
|
3SNL
 
 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | 分子名称: | 6-chloro-3,4-dimethyl-1-(3-methylpyridin-4-yl)-8-(trifluoromethyl)imidazo[1,5-a]quinoxaline, MAGNESIUM ION, ZINC ION, ... | | 著者 | Malamas, M.S, Ni, Y, Erdei, J, Stange, H, Schindler, R, Lankau, H.-J, Grunwald, C, Fan, K.Y, Parris, K.D, Langen, B, Egerland, U, Hage, T, Marquis, K.L, Grauer, S, Brennan, J, Navarra, R, Graf, R, Harrison, B.L, Robichaud, A, Kronbach, T, Pangalos, M, Hofgen, N, Brandon, N.J. | | 登録日 | 2011-06-29 | | 公開日 | 2011-10-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
3SNI
 
 | | Highly Potent, Selective, and Orally Active Phosphodiestarase 10A Inhibitors | | 分子名称: | 2-methoxy-6,7-dimethyl-9-(4-methylpyridin-3-yl)imidazo[1,5-a]pyrido[3,2-e]pyrazine, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Malamas, M.S, Ni, Y, Erdei, J, Stange, H, Schindler, R, Lankau, H.-J, Grunwald, C, Fan, K.Y, Parris, K.D, Langen, B, Egerland, U, Hage, T, Marquis, K.L, Grauer, S, Brennan, J, Navarra, R, Graf, R, Harrison, B.L, Robichaud, A, Kronbach, T, Pangalos, M, Hofgen, N, Brandon, N.J. | | 登録日 | 2011-06-29 | | 公開日 | 2011-10-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Highly Potent, Selective, and Orally Active Phosphodiesterase 10A Inhibitors.
J.Med.Chem., 54, 2011
|
|
7XPM
 
 | | Ancestral ADH WT | | 分子名称: | 1,2-ETHANEDIOL, A64 | | 著者 | Chen, X.Y, Xu, G.C, Ni, Y. | | 登録日 | 2022-05-04 | | 公開日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of a versatile ancestral ADH with high activity and thermostability
To Be Published
|
|
7YMU
 
 | | Structure of Alcohol dehydrogenase from [Candida] glabrata | | 分子名称: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sun, Z.W, Liu, Y.F, Xu, G.C, Ni, Y. | | 登録日 | 2022-07-29 | | 公開日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Rationla design of CgADH from candida glarata for asymmetric reduction of azacycolne.
To Be Published
|
|
7YMB
 
 | |
7YII
 
 | |
1EQ2
 
 | | THE CRYSTAL STRUCTURE OF ADP-L-GLYCERO-D-MANNOHEPTOSE 6-EPIMERASE | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, ADP-L-GLYCERO-D-MANNOHEPTOSE 6-EPIMERASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Deacon, A.M, Ni, Y.S, Coleman Jr, W.G, Ealick, S.E. | | 登録日 | 2000-03-31 | | 公開日 | 2000-11-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The crystal structure of ADP-L-glycero-D-mannoheptose 6-epimerase: catalysis with a twist.
Structure Fold.Des., 8, 2000
|
|
7V97
 
 | | Arsenic-bound p53 DNA-binding domain mutant V272M | | 分子名称: | ARSENIC, Cellular tumor antigen p53, ZINC ION | | 著者 | Lu, M, Xing, Y.F, Wang, Z.Y, Ni, Y, Song, H.X. | | 登録日 | 2021-08-24 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Diverse rescue potencies of p53 mutations to ATO are predetermined by intrinsic mutational properties.
Sci Transl Med, 15, 2023
|
|
6V97
 
 | | Kindlin-3 double deletion mutant short form | | 分子名称: | Fermitin family homolog 3 | | 著者 | Xu, Z, Zhang, T.L, Xu, Z, Sun, J.J, Ding, J.P, Ma, Y.Q. | | 登録日 | 2019-12-13 | | 公開日 | 2020-07-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.381 Å) | | 主引用文献 | Structure basis of the FERM domain of kindlin-3 in supporting integrin alpha IIb beta 3 activation in platelets.
Blood Adv, 4, 2020
|
|
6V9G
 
 | | Kindlin-3 double deletion mutant long form | | 分子名称: | Fermitin family homolog 3 | | 著者 | Xu, Z, Zhang, T.L, Xu, Z, Sun, J.J, Ding, J.P, Ma, Y.Q. | | 登録日 | 2019-12-13 | | 公開日 | 2020-07-22 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure basis of the FERM domain of kindlin-3 in supporting integrin alpha IIb beta 3 activation in platelets.
Blood Adv, 4, 2020
|
|
3P5B
 
 | | The structure of the LDLR/PCSK9 complex reveals the receptor in an extended conformation | | 分子名称: | CALCIUM ION, Low density lipoprotein receptor variant, Proprotein convertase subtilisin/kexin type 9 | | 著者 | Lo Surdo, P, Bottomley, M.J, Calzetta, A, Settembre, E.C, Cirillo, A, Pandit, S, Ni, Y, Hubbard, B, Sitlani, A, Carfi, A. | | 登録日 | 2010-10-08 | | 公開日 | 2011-10-26 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Mechanistic implications for LDL receptor degradation from the PCSK9/LDLR structure at neutral pH.
Embo Rep., 12, 2011
|
|
