7NA5
 
 | | Structure of the H2DB-TCR ternary complex with HSF2 melanoma neoantigen | | Descriptor: | 47BE7 TCR alpha chain, 47BE7 TCR beta chain, Beta-2-microglobulin, ... | | Authors: | Patskovsky, Y, Finnigan, J, Patskovska, L, Newman, J, Bhardwaj, N, Krogsgaard, M. | | Deposit date: | 2021-06-19 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the TCR-H2DB ternary complex with melanoma HSF2 neoantigen YGFRNVVHI
To be Published
|
|
7MY1
 
 | | Sy-CrtE structure with IPP, N-term His-tag | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CHLORIDE ION, Geranylgeranyl pyrophosphate synthase, ... | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
7MXZ
 
 | | Sy-CrtE apo structure | | Descriptor: | CHLORIDE ION, Geranylgeranyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
7MY7
 
 | | Se-CrtE N-term His-tag structure | | Descriptor: | Farnesyl-diphosphate synthase | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-20 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
7MY6
 
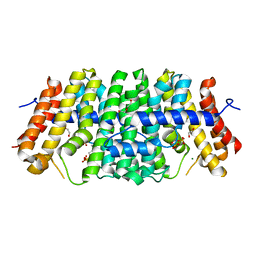 | | Se-CrtE C-term His-tag with IPP added | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
7RG7
 
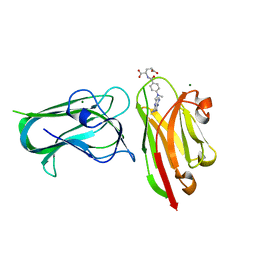 | | Crystal structure of nanoclamp8:VHH in complex with MTX | | Descriptor: | MAGNESIUM ION, METHOTREXATE, nano CLostridial Antibody Mimetic Protein 8 VHH | | Authors: | Guo, Z, Peat, T, Newman, J, Alexandrov, K. | | Deposit date: | 2021-07-14 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design of a methotrexate-controlled chemical dimerization system and its use in bio-electronic devices.
Nat Commun, 12, 2021
|
|
7SWS
 
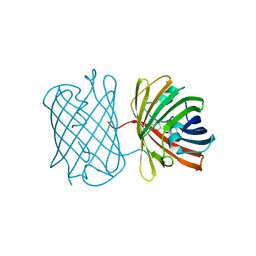 | | Crystal structure of the chromoprotein amilCP | | Descriptor: | BROMIDE ION, CHLORIDE ION, Chromoprotein amilCP | | Authors: | Caputo, A.T, Newman, J, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SWT
 
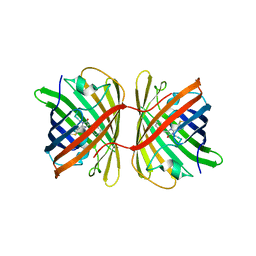 | | Crystal structure of the chromoprotein eforRED | | Descriptor: | Chromoprotein eforRED | | Authors: | Caputo, A.T, Newman, J, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SWR
 
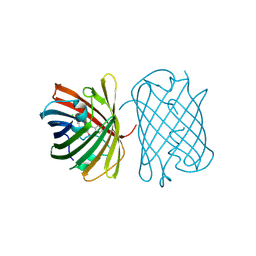 | | Crystal structure of the chromoprotein gfasPurple | | Descriptor: | CHLORIDE ION, Chromoprotein gfasPurple | | Authors: | Caputo, A.T, Newman, J, Peat, T.S, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.388 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SWU
 
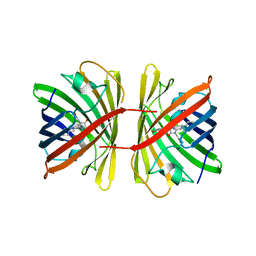 | | Crystal structure of the chromoprotein spisPINK | | Descriptor: | Chromoprotein spisPINK | | Authors: | Caputo, A.T, Newman, J, Scott, C, Ahmed, H. | | Deposit date: | 2021-11-21 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.444 Å) | | Cite: | Over the rainbow: structural characterization of the chromoproteins gfasPurple, amilCP, spisPink and eforRed.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7K09
 
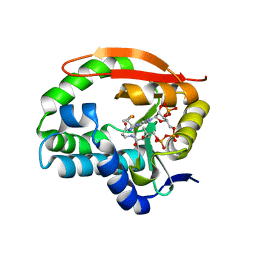 | | Puromycin N-acetyltransferase in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Puromycin N-acetyltransferase | | Authors: | Caputo, A.T, Newman, J, Adams, T.E, Peat, T.S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Structure-guided selection of puromycin N-acetyltransferase mutants with enhanced selection stringency for deriving mammalian cell lines expressing recombinant proteins.
Sci Rep, 11, 2021
|
|
7K0A
 
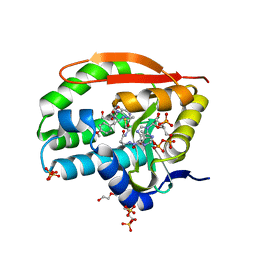 | | Puromycin N-acetyltransferase in complex with acetylated puromycin and CoA | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Caputo, A.T, Newman, J, Adams, T.E. | | Deposit date: | 2020-09-03 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided selection of puromycin N-acetyltransferase mutants with enhanced selection stringency for deriving mammalian cell lines expressing recombinant proteins.
Sci Rep, 11, 2021
|
|
4LH8
 
 | | Triazine hydrolase from Arthobacter aurescens modified for maximum expression in E.coli | | Descriptor: | Triazine hydrolase, ZINC ION | | Authors: | Jackson, C.J, Coppin, C.W, Alexandrov, A, Wilding, M, Liu, J.-W, Ubels, J, Paks, M, Carr, P.D, Newman, J, Russell, R.J, Field, M, Weik, M, Oakeshott, J.G, Scott, C. | | Deposit date: | 2013-07-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 300-Fold increase in production of the Zn2+-dependent dechlorinase TrzN in soluble form via apoenzyme stabilization.
Appl.Environ.Microbiol., 80, 2014
|
|
4L9X
 
 | | Triazine hydrolase from Arthobacter aurescens modified for maximum expression in E.coli | | Descriptor: | ACETATE ION, Triazine hydrolase | | Authors: | Jackson, C.J, Coppin, C.W, Alexandrov, A, Wilding, M, Liu, J.-W, Ubels, J, Paks, M, Carr, P.D, Newman, J, Russell, R.J, Field, M, Weik, M, Oakeshott, J.G, Scott, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 300-Fold increase in production of the Zn2+-dependent dechlorinase TrzN in soluble form via apoenzyme stabilization.
Appl.Environ.Microbiol., 80, 2014
|
|
6C62
 
 | | An unexpected vestigial protein complex reveals the evolutionary origins of an s-triazine catabolic enzyme. | | Descriptor: | AtzG, Biuret hydrolase, MAGNESIUM ION | | Authors: | Peat, T.S, Esquirol, L, Wilding, M, Liu, J.W, French, N.G, Hartley, C.J, Hideki, O, Easton, C.J, Newman, J, Scott, C. | | Deposit date: | 2018-01-17 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An unexpected vestigial protein complex reveals the evolutionary origins of ans-triazine catabolic enzyme.
J. Biol. Chem., 293, 2018
|
|
6C6G
 
 | | An unexpected vestigial protein complex reveals the evolutionary origins of an s-triazine catabolic enzyme. Inhibitor bound complex. | | Descriptor: | AtzG, Biuret hydrolase, CALCIUM ION | | Authors: | Peat, T.S, Esquirol, L, Wilding, M, Liu, J.W, French, N.G, Hartley, C.J, Hideki, O, Easton, C.J, Newman, J, Scott, C. | | Deposit date: | 2018-01-18 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An unexpected vestigial protein complex reveals the evolutionary origins of ans-triazine catabolic enzyme.
J. Biol. Chem., 293, 2018
|
|
4PTB
 
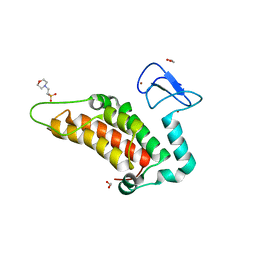 | | Crystal structure of human SP100 PHD-Bromodomain in the free state | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nuclear autoantigen Sp-100, ... | | Authors: | Tallant, C, Nunez-Alonso, G, Savitsky, P, Newman, J, Krojer, T, Szykowska, A, Burgess-Brown, N, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2014-03-10 | | Release date: | 2014-04-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of human SP100 PHD-Bromodomain in the free state
To be Published
|
|
6WEV
 
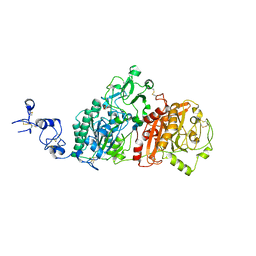 | | Crystal structures of human E-NPP 1: bound to N-{[1-(6,7-dimethoxy-5,8-dihydroquinazolin-4-yl)piperidin-4-yl]methyl}sulfuric diamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Peat, T.S, Dennis, M, Newman, J. | | Deposit date: | 2020-04-03 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human ENPP1 in apo and bound forms.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6WEU
 
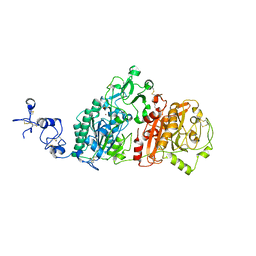 | | Crystal structures of human E-NPP 1: bound to adenosine-5'-thio-monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Peat, T.S, Dennis, M, Newman, J. | | Deposit date: | 2020-04-03 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of human ENPP1 in apo and bound forms.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6WFJ
 
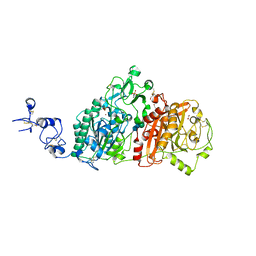 | | Crystal structures of human E-NPP 1: apo | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Peat, T.S, Dennis, M, Newman, J. | | Deposit date: | 2020-04-03 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human ENPP1 in apo and bound forms.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
2WZR
 
 | | The Structure of Foot and Mouth Disease Virus Serotype SAT1 | | Descriptor: | POLYPROTEIN | | Authors: | Adams, P, Lea, S, Newman, J, Blakemore, W, King, A, Stuart, D, Fry, E. | | Deposit date: | 2009-12-02 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Foot-and-Mouth Disease Virus Serotype Sat1.
To be Published
|
|
6AZN
 
 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Putative amidase | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
6AZQ
 
 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | CALCIUM ION, Putative amidase, dicarbonimidic diamide | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
6BJU
 
 | |
5G4Z
 
 | | Structural basis for carboxylic acid recognition by a Cache chemosensory domain. | | Descriptor: | Methyl-accepting chemotaxis sensory transducer with Cache sensor, TRIETHYLENE GLYCOL, UNKNOWN LIGAND | | Authors: | Brewster, J, McKellar, J.L.O, Newman, J, Peat, T.S, Gerth, M.L. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for ligand recognition by a Cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae.
Sci Rep, 6, 2016
|
|
