4UWX
 
 | | Structure of liprin-alpha3 in complex with mDia1 Diaphanous- inhibitory domain | | Descriptor: | LIPRIN-ALPHA-3, NICKEL (II) ION, PROTEIN DIAPHANOUS HOMOLOG 1, ... | | Authors: | Brenig, J, de Boor, S, Knyphausen, P, Kuhlmann, N, Wroblowski, S, Baldus, L, Scislowski, L, Artz, O, Trauschies, P, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2014-08-15 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Basis for the Inhibitory Effect of Liprin-Alpha3 on Mouse Diaphanous 1 (Mdia1) Function.
J.Biol.Chem., 290, 2015
|
|
4IB5
 
 | | Structure of human protein kinase CK2 catalytic subunit in complex with a CK2beta-competitive cyclic peptide | | Descriptor: | CHLORIDE ION, CK2beta-derived cyclic peptide, Casein kinase II subunit alpha, ... | | Authors: | Raaf, J, Guerra, B, Neundorf, I, Bopp, B, Issinger, O.-G, Jose, J, Pietsch, M, Niefind, K. | | Deposit date: | 2012-12-08 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | First structure of protein kinase CK2 catalytic subunit with an effective CK2 beta-competitive ligand
Acs Chem.Biol., 8, 2013
|
|
5FYQ
 
 | | Sirt2 in complex with a 13-mer trifluoroacetylated Ran peptide | | Descriptor: | NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2, RAN AA 31-43, SULFATE ION, ... | | Authors: | Knyphausen, P, de Boor, S, Scislowski, L, Extra, A, Baldus, L, Schacherl, M, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights Into Lysine-Deacetylation of Natively Folded Substrate Proteins by Sirtuins.
J.Biol.Chem., 291, 2016
|
|
6HG7
 
 | | Crystal structure of a collagen II fragment containing the binding site of PEDF and COMP, (POG)4-LKG HRG FTG LQG-POG(4) | | Descriptor: | Collagen alpha-1(II) chain, SULFATE ION | | Authors: | Gebauer, J.M, Koehler, A, Dietmar, H, Gompert, M, Neundorf, I, Zaucke, F, Koch, M, Baumann, U. | | Deposit date: | 2018-08-22 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | COMP and TSP-4 interact specifically with the novel GXKGHR motif only found in fibrillar collagens.
Sci Rep, 8, 2018
|
|
7AT9
 
 | | Structure of protein kinase ck2 catalytic subunit (csnk2a2 gene product) in complex with the ATP-competitive inhibitor MB002 and the alphaD-pocket ligand 3,4-dichlorophenethylamine | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dichlorophenyl)ethanamine, 3-(4,5,6,7-tetrabromo-1H-benzotriazol-1-yl)propan-1-ol, ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-29 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
7AT5
 
 | | Structure of protein kinase ck2 catalytic subunit (csnk2a1 gene product) in complex with the bivalent inhibitor KN2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dichlorophenyl)ethanamine, Casein kinase II subunit alpha, ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-29 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
7ATV
 
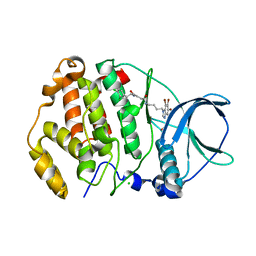 | | Structure of protein kinase ck2 catalytic subunit (csnk2a2 gene product) in complex with the bivalent inhibitor KN2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Casein kinase II subunit alpha', ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-31 | | Release date: | 2021-08-04 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
5A0S
 
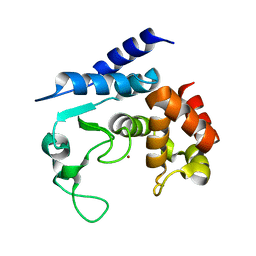 | | Apo-structure of metalloprotease Zmp1 variant E143A from Clostridium difficile | | Descriptor: | ZINC ION, ZINC METALLOPROTEASE ZMP1 | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-22 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
5A0R
 
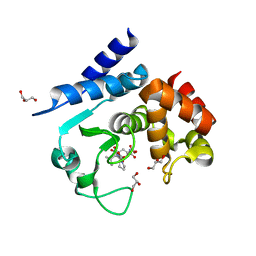 | | Product peptide-bound structure of metalloprotease Zmp1 variant E143A from Clostridium difficile | | Descriptor: | GLYCEROL, PRODUCT PEPTIDE, ZINC ION, ... | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-22 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
5A0P
 
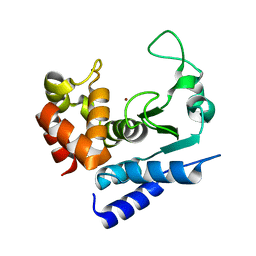 | | Apo-structure of metalloprotease Zmp1 from Clostridium difficile | | Descriptor: | ZINC ION, ZINC METALLOPROTEASE ZMP1 | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-22 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
5A0X
 
 | | Substrate peptide-bound structure of metalloprotease Zmp1 variant E143AY178F from Clostridium difficile | | Descriptor: | SUBSTRATE PEPTIDE, ZINC ION, ZINC METALLOPROTEASE ZMP1 | | Authors: | Schacherl, M, Pichlo, C, Neundorf, I, Baumann, U. | | Deposit date: | 2015-04-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Proline-Proline Peptide Bond Specificity of the Metalloprotease Zmp1 Implicated in Motility of Clostridium Difficile.
Structure, 23, 2015
|
|
6SYJ
 
 | |
2RND
 
 | |
2RMY
 
 | |
