4UYN
 
 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[5-(1H-benzimidazol-2-ylsulfanyl)furan-2-yl]-8-hydroxy-5,6,7,9-tetrahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Carry, J.C, Clerc, F, Minoux, H, Schio, L, Mauger, J, Nair, A, Parmantier, E, Lemoigne, R, Delorme, C, Nicolas, J.P, Krick, A, Abecassis, P.Y, Crocq-Stuerga, V, Pouzieux, S, Delarbre, L, Maignan, S, Bertrand, T, Bjergarde, K, Ma, N, Lachaud, S, Guizani, H, Lebel, R, Doerflinger, G, Monget, S, Perron, S, Gasse, F, Angouillant-Boniface, O, Filoche-Romme, B, Murer, M, Gontier, S, Prevost, C, Monteiro, M.L, Combeau, C. | | Deposit date: | 2014-09-02 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sar156497, an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
3ZT7
 
 | | GlgE isoform 1 from Streptomyces coelicolor with beta-cyclodextrin and maltose bound | | Descriptor: | Cycloheptakis-(1-4)-(alpha-D-glucopyranose), PUTATIVE GLUCANOHYDROLASE PEP1A, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rejzek, M, Fairhurst, S.A, Nair, A, Bruton, C.J, Field, R.A, Chater, K.F, Lawson, D.M, Bornemann, S. | | Deposit date: | 2011-07-01 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a Streptomyces Maltosyltransferase Glge: A Homologue of a Genetically Validated Anti-Tuberculosis Target.
J.Biol.Chem., 286, 2011
|
|
3ZT5
 
 | | GlgE isoform 1 from Streptomyces coelicolor with maltose bound | | Descriptor: | PUTATIVE GLUCANOHYDROLASE PEP1A, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rejzek, M, Fairhurst, S.A, Nair, A, Bruton, C.J, Field, R.A, Chater, K.F, Lawson, D.M, Bornemann, S. | | Deposit date: | 2011-07-01 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of a Streptomyces Maltosyltransferase Glge: A Homologue of a Genetically Validated Anti-Tuberculosis Target.
J.Biol.Chem., 286, 2011
|
|
3ZT6
 
 | | GlgE isoform 1 from Streptomyces coelicolor with alpha-cyclodextrin and maltose bound | | Descriptor: | Cyclohexakis-(1-4)-(alpha-D-glucopyranose), PUTATIVE GLUCANOHYDROLASE PEP1A, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rejzek, M, Fairhurst, S.A, Nair, A, Bruton, C.J, Field, R.A, Chater, K.F, Lawson, D.M, Bornemann, S. | | Deposit date: | 2011-07-01 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of a Streptomyces Maltosyltransferase Glge: A Homologue of a Genetically Validated Anti-Tuberculosis Target.
J.Biol.Chem., 286, 2011
|
|
3ZST
 
 | | GlgE isoform 1 from Streptomyces coelicolor with alpha-cyclodextrin bound | | Descriptor: | 1,2-ETHANEDIOL, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), PUTATIVE GLUCANOHYDROLASE PEP1A GLGE ISOFORM 1 | | Authors: | Syson, K, Stevenson, C.E.M, Rejzek, M, Fairhurst, S.A, Nair, A, Bruton, C.J, Field, R.A, Chater, K.F, Lawson, D.M, Bornemann, S. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a Streptomyces Maltosyltransferase Glge: A Homologue of a Genetically Validated Anti-Tuberculosis Target.
J.Biol.Chem., 286, 2011
|
|
4XYC
 
 | | NANOMOLAR INHIBITORS OF MYCOBACTERIUM TUBERCULOSIS GLUTAMINE SYNTHETASE 1: SYNTHESIS, BIOLOGICAL EVALUATION AND X-RAY CRYSTALLOGRAPHIC STUDIES | | Descriptor: | 9-phenyl-4H-imidazo[1,2-a]indeno[1,2-e]pyrazin-4-one, Glutamine synthetase 1 | | Authors: | Couturier, C, Silve, S, Morales, R, Ppessegue, B, Llopart, S, Nair, A, Bauer, A, Scheiper, B, poeverlein, c, Ganzhorn, A, Lagrange, S, Bacque, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Nanomolar inhibitors of Mycobacterium tuberculosis glutamine synthetase 1: Synthesis, biological evaluation and X-ray crystallographic studies.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
3ZSS
 
 | | Apo form of GlgE isoform 1 from Streptomyces coelicolor | | Descriptor: | PUTATIVE GLUCANOHYDROLASE PEP1A | | Authors: | Syson, K, Stevenson, C.E.M, Rejzek, M, Fairhurst, S.A, Nair, A, Bruton, C.J, Field, R.A, Chater, K.F, Lawson, D.M, Bornemann, S. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Streptomyces Maltosyltransferase Glge: A Homologue of a Genetically Validated Anti-Tuberculosis Target.
J.Biol.Chem., 286, 2011
|
|
1BC6
 
 | | 7-FE FERREDOXIN FROM BACILLUS SCHLEGELII, NMR, 20 STRUCTURES | | Descriptor: | 7-FE FERREDOXIN, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Aono, S, Bentrop, D, Bertini, I, Donaire, A, Luchinat, C, Niikura, Y, Rosato, A. | | Deposit date: | 1998-05-05 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized Fe7S8 ferredoxin from the thermophilic bacterium Bacillus schlegelii by 1H NMR spectroscopy.
Biochemistry, 37, 1998
|
|
1BD6
 
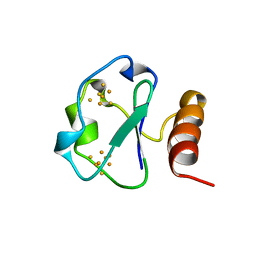 | | 7-FE FERREDOXIN FROM BACILLUS SCHLEGELII, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | 7-FE FERREDOXIN, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Aono, S, Bentrop, D, Bertini, I, Donaire, A, Luchinat, C, Niikura, Y, Rosato, A. | | Deposit date: | 1998-05-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized Fe7S8 ferredoxin from the thermophilic bacterium Bacillus schlegelii by 1H NMR spectroscopy.
Biochemistry, 37, 1998
|
|
1KQV
 
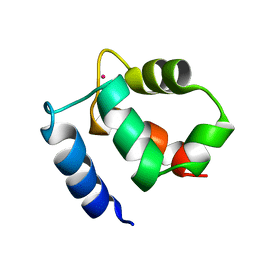 | | Family of NMR Solution Structures of Ca Ln Calbindin D9K | | Descriptor: | LANTHANUM (III) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN | | Authors: | Bertini, I, Donaire, A, Jimenez, B, Luchinat, C, Parigi, G, Piccioli, M, Poggi, L. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-based versus classical constraints: an analysis of the solution structure of Ca Ln calbindin D9k.
J.Biomol.NMR, 21, 2001
|
|
1KSM
 
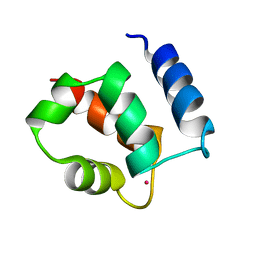 | | AVERAGE NMR SOLUTION STRUCTURE OF CA LN CALBINDIN D9K | | Descriptor: | LANTHANUM (III) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN | | Authors: | Bertini, I, Donaire, A, Luchinat, C, Piccioli, M, Poggi, L, Parigi, G, Jimenez, B. | | Deposit date: | 2002-01-14 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-based versus classical constraints: an analysis of the solution structure of Ca Ln calbindin D9k.
J.Biomol.NMR, 21, 2001
|
|
1CLF
 
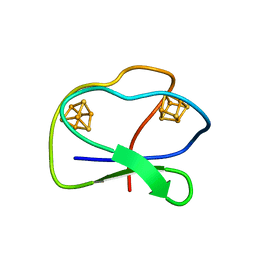 | | CLOSTRIDIUM PASTEURIANUM FERREDOXIN | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Bertini, I, Donaire, A, Feinberg, B.A, Luchinat, C, Piccioli, M, Yuan, H. | | Deposit date: | 1995-06-21 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized 2[4Fe-4S] ferredoxin from Clostridium pasteurianum.
Eur.J.Biochem., 232, 1995
|
|
2LLN
 
 | |
2JT5
 
 | | solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of n-hydroxy-2-[n-(2-hydroxyethyl)biphenyl-4-sulfonamide] hydroxamic acid (MLC88) | | Descriptor: | CALCIUM ION, N~2~-(biphenyl-4-ylsulfonyl)-N-hydroxy-N~2~-(2-hydroxyethyl)glycinamide, Stromelysin-1, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-07-20 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
2JNP
 
 | | Solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of N-isobutyl-N-[4-methoxyphenylsulfonyl]glycyl hydroxamic acid (NNGH) | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-3, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-01-30 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
2JT6
 
 | | Solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of 3-4'-cyanobyphenyl-4-yloxy)-n-hdydroxypropionamide (MMP-3 inhibitor VII) | | Descriptor: | 3-[(4'-cyanobiphenyl-4-yl)oxy]-N-hydroxypropanamide, CALCIUM ION, Stromelysin-1, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-07-23 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
6WBS
 
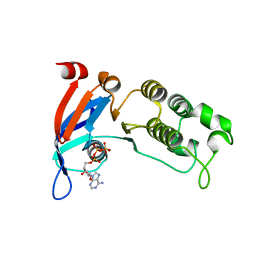 | | Human CFTR first nucleotide binding domain with dF508/V510D | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Simon, K.S, Kothe, M, Hilbert, B, Batchelor, J.D, Hurlbut, G.D. | | Deposit date: | 2020-03-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Determining the Molecular Mechanism of Suppressor Mutation V510D and the Contribution of Helical Unraveling to the dF508-CFTR Defect
To Be Published
|
|
4UZD
 
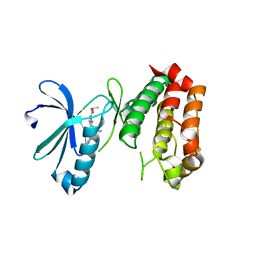 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[3-(1H-benzimidazol-2-yloxy)phenyl]-8-oxo-4,5,6,7,8,9-hexahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Pouzieux, S, Delarbre, L, Crenne, J.Y. | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Sar156497 an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
4UZH
 
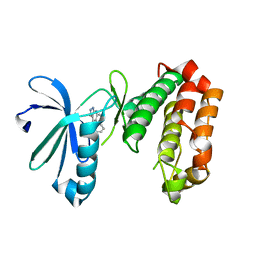 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | (4S)-4-(2-fluorophenyl)-2,4,6,7,8,9-hexahydro-5H-pyrazolo[3,4-b][1,7]naphthyridin-5-one, AURORA 2 KINASE DOMAIN | | Authors: | Pouzieux, S, Maignan, S, Crenne, J.Y. | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sar156497 an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
3O23
 
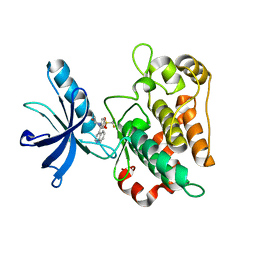 | |
