2F62
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.5 A resolution with (2-ETHYLPHENYL)METHANOL bound | | Descriptor: | (2-ETHYLPHENYL)METHANOL, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2F2T
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.7 A resolution with 5-Aminoisoquinoline bound | | Descriptor: | GLYCEROL, ISOQUINOLIN-5-AMINE, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-17 | | Release date: | 2005-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2F64
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.6 A resolution with 1-METHYLQUINOLIN-2(1H)-ONE bound | | Descriptor: | 1-METHYLQUINOLIN-2(1H)-ONE, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
4WGJ
 
 | |
4WI1
 
 | |
4XI8
 
 | |
4Y0V
 
 | |
4YET
 
 | | X-ray crystal structure of superoxide dismutase from Babesia bovis solved by Sulfur SAD | | Descriptor: | FE (III) ION, Superoxide dismutase | | Authors: | Fairman, J.W, Clifton, M.C, Abendroth, J, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2015-02-24 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Iron superoxide dismutases in eukaryotic pathogens: new insights from Apicomplexa and Trypanosoma structures.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4YLG
 
 | |
4ZDQ
 
 | |
5BT9
 
 | |
5ELN
 
 | |
5ELO
 
 | |
1X9G
 
 | |
1XN4
 
 | |
1YZV
 
 | |
2A0K
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.8 A resolution | | Descriptor: | GLYCEROL, Nucleoside 2-deoxyribosyltransferase, SULFATE ION | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-06-16 | | Release date: | 2005-07-26 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2A0M
 
 | |
2AR1
 
 | |
4IX8
 
 | |
4P9A
 
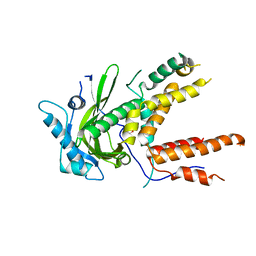 | | X-ray Crystal Structure of PA protein from Influenza strain H7N9 | | Descriptor: | GLYCEROL, Polymerase PA | | Authors: | Fairman, J.W, Moen, S.O, Clifton, M.C, Edwards, T.E, Lorimer, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1999 Å) | | Cite: | Structural analysis of H1N1 and H7N9 influenza A virus PA in the absence of PB1.
Sci Rep, 4, 2014
|
|
6MB0
 
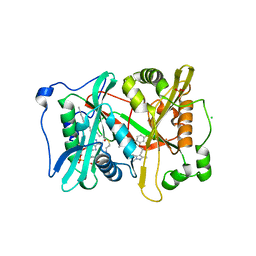 | |
6MB1
 
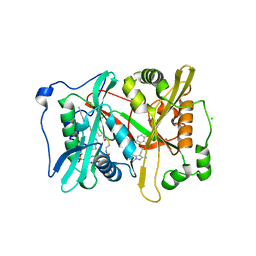 | | Crystal structure of N-myristoyl transferase (NMT) from Plasmodium vivax in complex with inhibitor IMP-1002 | | Descriptor: | 1,2-ETHANEDIOL, 1-(5-{4-fluoro-2-[2-(1,3,5-trimethyl-1H-pyrazol-4-yl)ethoxy]phenyl}-1-methyl-1H-indazol-3-yl)-N,N-dimethylmethanamine, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-08-29 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Identification of Resistance Breaking Antimalarial N‐Myristoyltransferase Inhibitors.
Cell Chem Biol, 26, 2019
|
|
6MFK
 
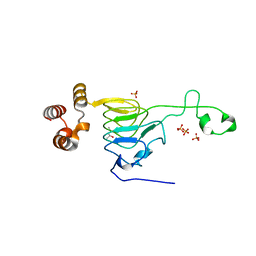 | |
4Q6U
 
 | |
