379D
 
 | | THE STRUCTURAL BASIS OF HAMMERHEAD RIBOZYME SELF-CLEAVAGE | | Descriptor: | COBALT (II) ION, RNA (5'-R(*GP*GP*CP*CP*GP*AP*AP*AP*CP*UP*CP*GP*UP*AP*AP*GP*A P*GP*UP*CP*AP*CP*CP*AP*C)-3'), RNA (5'-R(*GP*UP*GP*GP*UP*CP*UP*GP*AP*UP*GP*AP*GP*GP*CP*C)-3') | | Authors: | Murray, J.B, Terwey, D.P, Maloney, L, Karpeisky, A, Usman, N, Beigelman, L, Scott, W.G. | | Deposit date: | 1998-02-05 | | Release date: | 1998-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis of hammerhead ribozyme self-cleavage.
Cell(Cambridge,Mass.), 92, 1998
|
|
488D
 
 | | CATALYTIC RNA ENZYME-PRODUCT COMPLEX | | Descriptor: | CADMIUM ION, FIRST RNA FRAGMENT OF CLEAVED SUBSTRATE, RNA RIBOZYME STRAND, ... | | Authors: | Murray, J.B, Szoke, H, Szoke, A, Scott, W.G. | | Deposit date: | 2000-02-25 | | Release date: | 2000-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Capture and visualization of a catalytic RNA enzyme-product complex using crystal lattice trapping and X-ray holographic reconstruction.
Mol.Cell, 5, 2000
|
|
2F4V
 
 | | 30S ribosome + designer antibiotic | | Descriptor: | (2R)-4-AMINO-N-{(1R,2S,3R,4R,5S)-5-AMINO-2-{2-[(2-AMINOETHYL)AMINO]ETHOXY}-4-[(2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-3-HYDROXYCYCLOHEXYL}-2-HYDROXYBUTANAMIDE, (2S,4S,4AR,5AS,6S,11R,11AS,12R,12AR)-7-CHLORO-4-(DIMETHYLAMINO)-6,10,11,12-TETRAHYDROXY-1,3-DIOXO-1,2,3,4,4A,5,5A,6,11,11A,12,12A-DODECAHYDROTETRACENE-2-CARBOXAMIDE, 16S ribosomal RNA, ... | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosome aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
2F4S
 
 | | A-site RNA in complex with neamine | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 5'-R(P*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosomal aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
2F4T
 
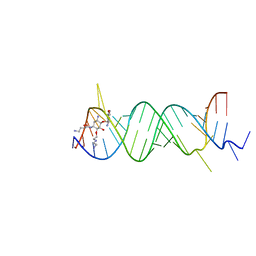 | | Asite RNA + designer antibiotic | | Descriptor: | (2R)-4-AMINO-N-{(1R,2S,3R,4R,5S)-5-AMINO-2-{2-[(2-AMINOETHYL)AMINO]ETHOXY}-4-[(2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-3-HYDROXYCYCLOHEXYL}-2-HYDROXYBUTANAMIDE, 5'-R(*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3', 5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosomal aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
2F4U
 
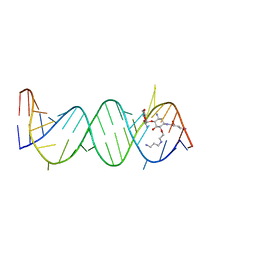 | | Asite RNA + designer antibiotic | | Descriptor: | (2R)-4-AMINO-N-((1R,2S,3R,4R,5S)-5-AMINO-4-[(2-AMINO-2-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-{2-[(3-AMINOPROPYL)AMINO]ETHOXY}-3-HYDROXYCYCLOHEXYL)-2-HYDROXYBUTANAMIDE, 5'-R(*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosomal aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
3KAD
 
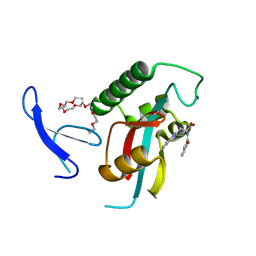 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 3-(1H-benzimidazol-2-yl)-N-(3-phenylpropanoyl)-D-alanine, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KAB
 
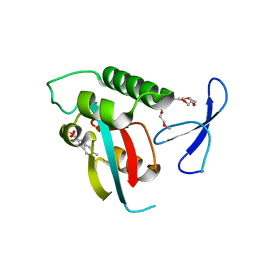 | | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors | | Descriptor: | 6-methyl-1H-indole-2-carboxylic acid, DODECAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Baker, L.M, Dokurno, P, Robinson, D.A, Surgenor, A.E, Murray, J.B, Potter, A.J, Moore, J.D. | | Deposit date: | 2009-10-19 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure-guided design of alpha-amino acid-derived Pin1 inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6MSF
 
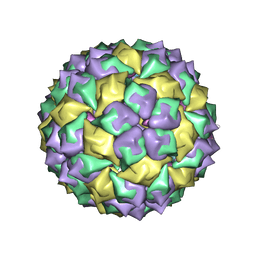 | | F6 APTAMER MS2 COAT PROTEIN COMPLEX | | Descriptor: | PROTEIN (MS2 PROTEIN CAPSID), RNA (5'-R(*CP*AP*GP*UP*CP*AP*CP*UP*GP*G)-3'), RNA (5'-R(*CP*CP*AP*CP*AP*GP*UP*CP*AP*CP*UP*GP*GP*G)-3') | | Authors: | Convery, M.A, Rowsell, S, Stonehouse, N.J, Ellington, A.D, Hirao, I, Murray, J.B, Peabody, D.S, Phillips, S.E.V, Stockley, P.G. | | Deposit date: | 1998-01-06 | | Release date: | 1998-07-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an RNA aptamer-protein complex at 2.8 A resolution.
Nat.Struct.Biol., 5, 1998
|
|
6TMZ
 
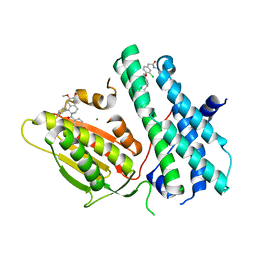 | | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures | | Descriptor: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, [2,4-bis(oxidanyl)phenyl]-[(1~{R})-6,7-dimethoxy-1-pyridin-3-yl-3,4-dihydro-1~{H}-isoquinolin-2-yl]methanone, ... | | Authors: | Baker, L.M, Aimon, A, Murray, J.B, Surgenor, A.E, Matassova, N, Roughley, S.D, von Delft, F, Hubbard, R.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures
Commun Chem, 2020
|
|
6TN2
 
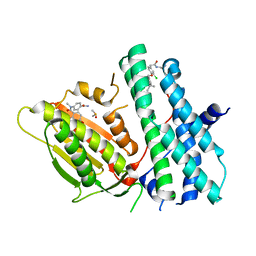 | | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, ... | | Authors: | Baker, L.M, Aimon, A, Murray, J.B, Surgenor, A.E, Matassova, N, Roughley, S.D, von Delft, F, Hubbard, R.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures
Commun Chem, 2020
|
|
6TN5
 
 | | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures | | Descriptor: | CHLORIDE ION, Heat shock protein HSP 90-alpha, ~{N}-(4-aminocarbonylphenyl)-~{N}-methyl-2,4-bis(oxidanyl)benzamide | | Authors: | Baker, L.M, Aimon, A, Murray, J.B, Surgenor, A.E, Matassova, N, Roughley, S.D, von Delft, F, Hubbard, R.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.165 Å) | | Cite: | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures
Commun Chem, 2020
|
|
3ODK
 
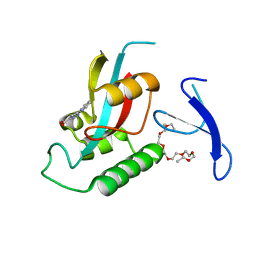 | | Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-pyridin-2-yl-1H-pyrazole-5-carboxylic acid, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Potter, A, Oldfield, V, Nunns, C, Fromont, C, Ray, S, Northfield, C.J, Bryant, C.J, Scrace, S.F, Robinson, D, Matossova, N, Baker, L, Dokurno, P, Surgenor, A.E, Davis, B.E, Richardson, C.M, Murray, J.B, Moore, J.D. | | Deposit date: | 2010-08-11 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5OOP
 
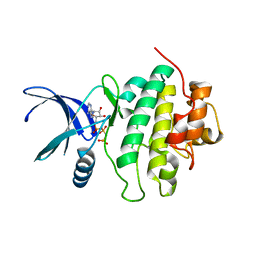 | | Structure of CHK1 10-pt. mutant complex with AMP-PNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OP5
 
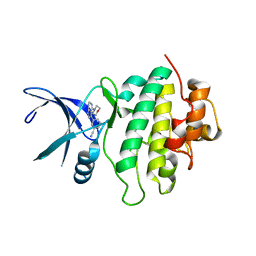 | | Structure of CHK1 10-pt. mutant complex with pyrrolopyrimidine LRRK2 inhibitor | | Descriptor: | 3-[4-(morpholin-4-yl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]benzonitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OQ6
 
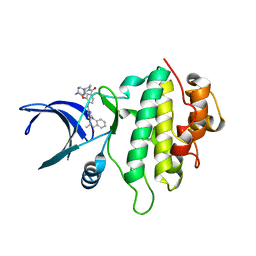 | | Structure of CHK1 12-pt. mutant complex with aminopyrimido-benzodiazepinone LRRK2 inhibitor | | Descriptor: | 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OOT
 
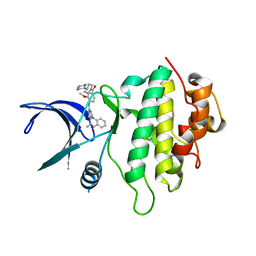 | | Structure of CHK1 10-pt. mutant complex with aminopyrimido-benzodiazepinone LRRK2 inhibitor | | Descriptor: | 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OP2
 
 | | Structure of CHK1 10-pt. mutant complex with arylbenzamide LRRK2 inhibitor | | Descriptor: | 5-(4-methylpiperazin-1-yl)-2-phenylmethoxy-~{N}-pyridin-3-yl-benzamide, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OPB
 
 | | Structure of CHK1 10-pt. mutant complex with indazole LRRK2 inhibitor | | Descriptor: | (2~{R},6~{S})-2,6-dimethyl-4-[6-[5-(1-methylcyclopropyl)oxy-1~{H}-indazol-3-yl]pyrimidin-4-yl]morpholine, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OQ8
 
 | | Structure of CHK1 12-pt. mutant complex with arylbenzamide LRRK2 inhibitor | | Descriptor: | 5-(4-methylpiperazin-1-yl)-2-phenylmethoxy-~{N}-pyridin-3-yl-benzamide, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OOR
 
 | | Structure of CHK1 10-pt. mutant complex with staurosporine | | Descriptor: | CHLORIDE ION, STAUROSPORINE, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OQ7
 
 | | Structure of CHK1 8-pt. mutant complex with arylbenzamide LRRK2 inhibitor | | Descriptor: | 5-(4-methylpiperazin-1-yl)-2-phenylmethoxy-~{N}-pyridin-3-yl-benzamide, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OPR
 
 | | Structure of CHK1 10-pt. mutant complex with aminopyridine LRRK2 inhibitor | | Descriptor: | 5-[4-(morpholin-4-ylmethyl)phenyl]-3-(1-propan-2-yl-1,2,3-triazol-4-yl)pyridin-2-amine, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OPV
 
 | | Structure of CHK1 10-pt. mutant complex with pyrrolopyridine LRRK2 inhibitor | | Descriptor: | 4-(3-methylphenyl)-6-[(1-methylpyrazol-3-yl)amino]-1~{H}-pyrrolo[2,3-b]pyridine-3-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
5OQ5
 
 | | Structure of CHK1 8-pt. mutant complex with aminopyrimido-benzodiazepinone LRRK2 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Williamson, D.S, Acheson-Dossang, P, Chen, I, Murray, J.B, Shaw, T, Surgenor, A.E. | | Deposit date: | 2017-08-10 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design of Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Crystallographic Surrogate Derived from Checkpoint Kinase 1 (CHK1).
J. Med. Chem., 60, 2017
|
|
