1Z5T
 
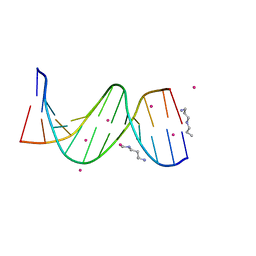 | | Crystal Structure of [d(CGCGAA(Z3dU)(Z3dU)CGCG)]2, Z3dU:5-(3-aminopropyl)-2'-deoxyuridine, in presence of thallium I. | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*(ZDU)P*(ZDU)P*CP*GP*CP*G)-3', SPERMINE, THALLIUM (I) ION | | Authors: | Moulaei, T, Maehigashi, T, Lountos, G.T, Komeda, S, Watkins, D, Stone, M.P, Marky, L.A, Li, J.S, Gold, B, Williams, L.D. | | Deposit date: | 2005-03-19 | | Release date: | 2005-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of B-DNA with cations tethered in the major groove.
Biochemistry, 44, 2005
|
|
2QSK
 
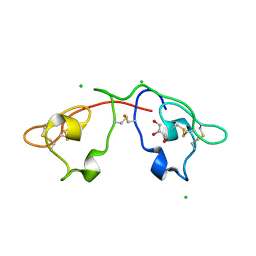 | | Atomic-resolution crystal structure of the Recombinant form of Scytovirin | | Descriptor: | CHLORIDE ION, GLYCEROL, scytovirin | | Authors: | Moulaei, T, Botos, I, Ziolkowska, N.E, Dauter, Z, Wlodawer, A. | | Deposit date: | 2007-07-31 | | Release date: | 2007-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic-resolution crystal structure of the antiviral lectin scytovirin.
Protein Sci., 16, 2007
|
|
2QT4
 
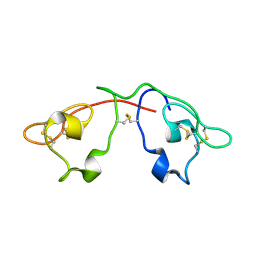 | | Atomic-resolution crystal structure of the natural form of Scytovirin | | Descriptor: | scytovirin | | Authors: | Moulaei, T, Botos, I, Ziolkowska, N.E, Dauter, Z, Wlodawer, A. | | Deposit date: | 2007-08-01 | | Release date: | 2007-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic-resolution crystal structure of the antiviral lectin scytovirin.
Protein Sci., 16, 2007
|
|
3LL1
 
 | |
3LKY
 
 | | Monomeric Griffithsin with a Single Gly-Ser Insertion | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Griffithsin, ... | | Authors: | Moulaei, T, Wlodawer, A. | | Deposit date: | 2010-01-28 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Monomerization of viral entry inhibitor griffithsin elucidates the relationship between multivalent binding to carbohydrates and anti-HIV activity.
Structure, 18, 2010
|
|
3LL2
 
 | |
3LL0
 
 | | Monomeric Griffithsin with two Gly-Ser Insertions | | Descriptor: | GLYCEROL, Griffithsin, SULFATE ION | | Authors: | Moulaei, T, Wlodawer, A. | | Deposit date: | 2010-01-28 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Monomerization of viral entry inhibitor griffithsin elucidates the relationship between multivalent binding to carbohydrates and anti-HIV activity.
Structure, 18, 2010
|
|
3GGI
 
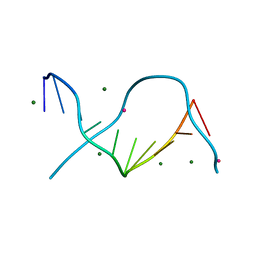 | | Locating monovalent cations in one turn of G/C rich B-DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*CP*CP*TP*GP*G) -3', MAGNESIUM ION, THALLIUM (I) ION | | Authors: | Maehigashi, T, Moulaei, T, Watkins, D, Komeda, S, Williams, L.D. | | Deposit date: | 2009-02-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Locating monovalent cations in one turn of G/C rich B-DNA
To be Published
|
|
3GGK
 
 | | Locating monovalent cations in one turn of G/C rich B-DNA | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*CP*CP*TP*GP*G) -3', MAGNESIUM ION, RUBIDIUM ION | | Authors: | Maehigashi, T, Moulaei, T, Watkins, D, Komeda, S, Williams, L.D. | | Deposit date: | 2009-02-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | Locating monovalent cations in one turn of G/C rich B-DNA
To be Published
|
|
5MLX
 
 | | Open loop conformation of PhaZ7 Y105E mutant | | Descriptor: | CHLORIDE ION, PHB depolymerase PhaZ7, SODIUM ION | | Authors: | Kellici, T, Mavromoustakos, T, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2016-12-08 | | Release date: | 2017-05-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure analysis, covalent docking, and molecular dynamics calculations reveal a conformational switch in PhaZ7 PHB depolymerase.
Proteins, 85, 2017
|
|
5MLY
 
 | | Closed loop conformation of PhaZ7 Y105E mutant | | Descriptor: | PHB depolymerase PhaZ7 | | Authors: | Kellici, T, Mavromoustakos, T, Jendrossek, D, Papageorgiou, A.C. | | Deposit date: | 2016-12-08 | | Release date: | 2017-05-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal structure analysis, covalent docking, and molecular dynamics calculations reveal a conformational switch in PhaZ7 PHB depolymerase.
Proteins, 85, 2017
|
|
4ZB4
 
 | | Spliceosome component | | Descriptor: | Pre-mRNA-processing factor 19 | | Authors: | Rocha de Moura, T, Pena, P. | | Deposit date: | 2015-04-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Spliceosome component
To Be Published
|
|
5EG7
 
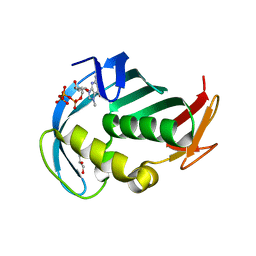 | | The cap binding site of influenza virus protein PB2 as a drug target | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, GLYCEROL, Polymerase basic protein 2 | | Authors: | Severin, C, Rocha de Moura, T, Liu, Y, Li, K, Zheng, X, Luo, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The cap-binding site of influenza virus protein PB2 as a drug target.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EG8
 
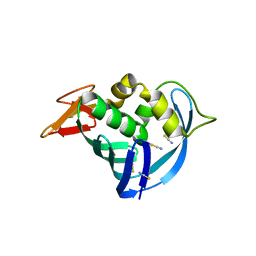 | | The cap binding site of influenza virus protein PB2 as a drug target | | Descriptor: | Polymerase basic protein 2, THIOCYANATE ION | | Authors: | Liu, Y, Zheng, X, Severin, C, Rocha de Moura, T, Li, K, Luo, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The cap-binding site of influenza virus protein PB2 as a drug target.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5EG9
 
 | | The cap binding site of influenza virus protein PB2 as a drug target | | Descriptor: | Polymerase basic protein 2 | | Authors: | Severin, C, Rocha de Moura, T, Liu, Y, Li, K, Zheng, X, Luo, M. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The cap-binding site of influenza virus protein PB2 as a drug target.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3HIH
 
 | |
3HIK
 
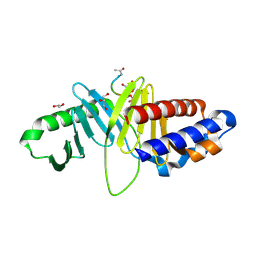 | | Structure of human Plk1-PBD in complex with PLHSpT | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pentamer phosphopeptide, ... | | Authors: | Wlodawer, A, Moulaei, T. | | Deposit date: | 2009-05-20 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and functional analyses of minimal phosphopeptides targeting the polo-box domain of polo-like kinase 1.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3U89
 
 | | Crystal structure of one turn of g/c rich b-dna revisited | | Descriptor: | 5'-D(*CP*CP*AP*GP*GP*CP*CP*TP*GP*G) -3', MAGNESIUM ION | | Authors: | Maehigashi, T, Woods, K.K, Moulaei, T, Komeda, S, Williams, L.D. | | Deposit date: | 2011-10-16 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | B-DNA structure is intrinsically polymorphic: even at the level of base pair positions.
Nucleic Acids Res., 40, 2012
|
|
2DYW
 
 | | A Backbone binding DNA complex | | Descriptor: | (6-AMINOHEXYLAMINE)(TRIAMMINE) PLATINUM(II) COMPLEX, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', SODIUM ION, ... | | Authors: | Komeda, S, Moulaei, T, Woods, K.K, Chikuma, M, Farrell, N.P, Williams, L.D. | | Deposit date: | 2006-09-18 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | A Third Mode of DNA Binding: Phosphate Clamps by a Polynuclear Platinum Complex
J.Am.Chem.Soc., 128, 2006
|
|
6UES
 
 | | Apo SAM-IV Riboswitch | | Descriptor: | RNA (119-MER) | | Authors: | Zhang, K, Li, S, Kappel, K, Pintilie, G, Su, Z, Mou, T, Schmid, M, Das, R, Chiu, W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 angstrom resolution.
Nat Commun, 10, 2019
|
|
6UET
 
 | | SAM-bound SAM-IV riboswitch | | Descriptor: | RNA (119-MER), S-ADENOSYLMETHIONINE | | Authors: | Zhang, K, Li, S, Kappel, K, Pintilie, G, Su, Z, Mou, T, Schmid, M, Das, R, Chiu, W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 angstrom resolution.
Nat Commun, 10, 2019
|
|
7RKA
 
 | |
5KHT
 
 | | Crystal structure of the N-terminal fragment of tropomyosin isoform Tpm1.1 at 1.5 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tropomyosin alpha-1 chain,General control protein GCN4 | | Authors: | Kostyukova, A.S, Krieger, I, Yoon, Y.-H, Tolkatchev, D, Samatey, F.A. | | Deposit date: | 2016-06-15 | | Release date: | 2017-06-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4964 Å) | | Cite: | Structural destabilization of tropomyosin induced by the cardiomyopathy-linked mutation R21H.
Protein Sci., 27, 2018
|
|
5Z9C
 
 | |
3U4X
 
 | | Crystal structure of a lectin from Camptosema pedicellatum seeds in complex with 5-bromo-4-chloro-3-indolyl-alpha-D-mannose | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Camptosema pedicellatum lectin (CPL), ... | | Authors: | Rocha, B.A.M, Teixeira, C.S, Moura, T.R, Silva, H.C, Pereira-Junior, F.N, Nagano, C.S, Delatorre, P, Cavada, B.S. | | Deposit date: | 2011-10-10 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of the lectin of Camptosema pedicellatum: implications of a conservative substitution at the hydrophobic subsite.
J.Biochem., 152, 2012
|
|
