1NRW
 
 | | The structure of a HALOACID DEHALOGENASE-LIKE HYDROLASE FROM B. SUBTILIS | | Descriptor: | CALCIUM ION, PHOSPHATE ION, hypothetical protein, ... | | Authors: | Cuff, M.E, Kim, Y, Zhang, R, Joachimiak, A, Collart, F, Quartey, P, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-25 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of a HALOACID DEHALOGENASE-LIKE HYDROLASE FROM B. SUBTILIS
To be Published
|
|
4GXT
 
 | | The crystal structure of a conserved functionally unknown protein from Anaerococcus prevotii DSM 20548 | | Descriptor: | GLYCEROL, SULFATE ION, a conserved functionally unknown protein | | Authors: | Tan, K, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-04 | | Release date: | 2012-10-03 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | The crystal structure of a conserved functionally unknown protein from Anaerococcus prevotii DSM 20548
To be Published
|
|
4H7N
 
 | |
4ML9
 
 | | Crystal Structure of Uncharacterized TIM Barrel Protein with the Conserved Phosphate Binding Site fromSebaldella termitidis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Holowicki, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Crystal Structure of Uncharacterized TIM Barrel Protein with the Conserved Phosphate Binding Site fromSebaldella termitidis
To be Published
|
|
4H2K
 
 | | Crystal structure of the catalytic domain of succinyl-diaminopimelate desuccinylase from Haemophilus influenzae | | Descriptor: | Succinyl-diaminopimelate desuccinylase, ZINC ION | | Authors: | Nocek, B, Jedrzejczak, R, Makowska-Grzyska, M, Starus, A, Holz, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-12 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The dimerization domain in DapE enzymes is required for catalysis.
Plos One, 9, 2014
|
|
4HB7
 
 | | The Structure of Dihydropteroate Synthase from Staphylococcus aureus subsp. aureus Mu50. | | Descriptor: | 1,2-ETHANEDIOL, Dihydropteroate synthase | | Authors: | Cuff, M.E, Holowicki, J, Jedrzejczak, R, Terwilliger, T.C, Rubin, E.J, Guinn, K, Baker, D, Ioerger, T.R, Sacchettini, J.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structure of Dihydropteroate Synthase from Staphylococcus aureus subsp. aureus Mu50.
TO BE PUBLISHED
|
|
4GZE
 
 | | Crystal structure of 6-phospho-beta-glucosidase from Lactobacillus plantarum (apo form) | | Descriptor: | 6-phospho-beta-glucosidase, CHLORIDE ION, GLYCEROL | | Authors: | Michalska, K, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-06 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HES
 
 | | Structure of a Beta-Lactamase Class A-like Protein from Veillonella parvula. | | Descriptor: | Beta-lactamase class A-like protein, FORMIC ACID, GLYCEROL, ... | | Authors: | Cuff, M.E, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-10-04 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Beta-Lactamase Class A-like Protein from Veillonella parvula.
TO BE PUBLISHED
|
|
4GYU
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 A121F mutant from Klebsiella pneumoniae | | Descriptor: | Beta-lactamase NDM-1, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 A121F mutant from Klebsiella pneumoniae
To be Published, 2012
|
|
4HL1
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1, Complexed with Cd and Ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase NDM-1, CADMIUM ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-10-15 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1, Complexed with Cd and Ampicillin
To be Published
|
|
4HNH
 
 | | The crystal structure of a short-chain dehydrogenases/reductase (wide type) from Veillonella parvula DSM 2008 in complex with NADP | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-10-19 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.576 Å) | | Cite: | The crystal structure of a short-chain dehydrogenases/reductase (wide type) from Veillonella parvula DSM 2008 in complex with NADP.
To be Published
|
|
4HKY
 
 | | New Delhi Metallo-beta-Lactamase-1, Complexed with Cd and Faropenem | | Descriptor: | (2R)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-5-[(2R)-oxolan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylic acid, (5R,6S)-6-(1-hydroxyethyl)-7-oxo-3-[(2R)-oxolan-2-yl]-4-thia-1-azabicyclo[3.2.0]hept-2-ene-2-carboxylic acid, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-10-15 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | New Delhi Metallo-beta-Lactamase-1, Complexed with Cd and Faropenem
To be Published
|
|
4I6V
 
 | | The crystal structure of an amidohydrolase 2 from Planctomyces limnophilus DSM 3776 | | Descriptor: | ACETATE ION, Amidohydrolase 2, GLYCEROL, ... | | Authors: | Fan, Y, Tan, K, Wu, R, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | The crystal structure of an amidohydrolase 2 from Planctomyces limnophilus DSM 3776
To be Published
|
|
4HYL
 
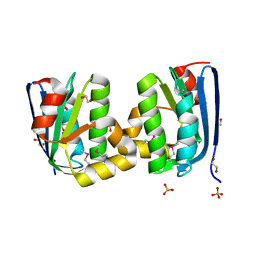 | | The crystal structure of an anti-sigma-factor antagonist from Haliangium ochraceum DSM 14365 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Stage II sporulation protein | | Authors: | Tan, K, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-13 | | Release date: | 2012-11-28 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The crystal structure of an anti-sigma-factor antagonist from Haliangium ochraceum DSM 14365
To be Published
|
|
4I4J
 
 | | The structure of SgcE10, the ACP-polyene thioesterase involved in C-1027 biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, ACP-polyene thioesterase, D(-)-TARTARIC ACID, ... | | Authors: | Kim, Y, Bigelow, L, Bearden, J, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Crystal Structure of Thioesterase SgcE10 Supporting Common Polyene Intermediates in 9- and 10-Membered Enediyne Core Biosynthesis.
Acs Omega, 2, 2017
|
|
4I19
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | ACETATE ION, Epoxide hydrolase, FORMIC ACID | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-05 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
1Q7H
 
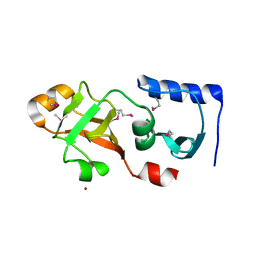 | | Structure of a Conserved PUA Domain Protein from Thermoplasma acidophilum | | Descriptor: | ZINC ION, conserved hypothetical protein | | Authors: | Cuff, M.E, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-18 | | Release date: | 2004-01-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a conserved hypothetical protein from T. acidophilum
TO BE PUBLISHED
|
|
1Q77
 
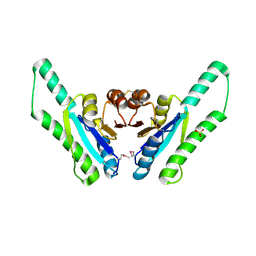 | | X-ray crystal structure of putative Universal Stress Protein from Aquifex aeolicus | | Descriptor: | Hypothetical protein AQ_178, SULFATE ION | | Authors: | Osipiuk, J, Zhou, M, Moy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-16 | | Release date: | 2003-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural homolog of Universal Stress Protein from Aquifex aeolicus
To be Published
|
|
4IQN
 
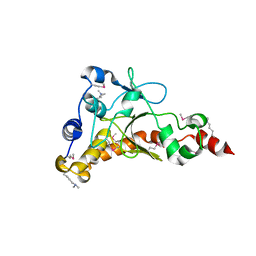 | | Crystal structure of uncharacterized protein from Salmonella enterica subsp. enterica serovar typhimurium str. 14028s | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative cytoplasmic protein, TETRAETHYLENE GLYCOL | | Authors: | Chang, C, Hatzos-Skintges, C, Adkins, J.N, Brown, R.N, Cort, J.R, Heffron, F, Nakayasu, E.S, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Program for the Characterization of Secreted Effector Proteins (PCSEP) | | Deposit date: | 2013-01-11 | | Release date: | 2013-01-23 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of uncharacterized protein from salmonella enterica subsp. enterica serovar typhimurium str. 14028s
To be Published
|
|
1Q8B
 
 | |
4J01
 
 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 29mer DNA target | | Descriptor: | DNA (29-MER), SULFATE ION, Transcription Factor HetR | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.246 Å) | | Cite: | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1R0U
 
 | | Crystal structure of ywiB protein from Bacillus subtilis | | Descriptor: | GLYCEROL, protein ywiB | | Authors: | Osipiuk, J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of ywiB protein from Bacillus subtilis
to be published
|
|
4IYI
 
 | | The crystal structure of a secreted protein EsxB (wild-type, C-term. His-tagged) from Bacillus anthracis str. Sterne | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.083 Å) | | Cite: | The crystal structure of a secreted protein EsxB (wild-type, C-term. His-tagged) from Bacillus anthracis str. Sterne
To be Published
|
|
4IYH
 
 | | The crystal structure of a secreted protein EsxB (SeMet-labeled, C-term. His-Tagged) from Bacillus anthracis str. Sterne | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | The crystal structure of a secreted protein EsxB (SeMet-labeled, C-term. His-Tagged) from Bacillus anthracis str. Sterne
To be Published
|
|
4OPE
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmH KS7 | | Descriptor: | NITRATE ION, NRPS/PKS | | Authors: | Osipiuk, J, Mack, J, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
