1DPU
 
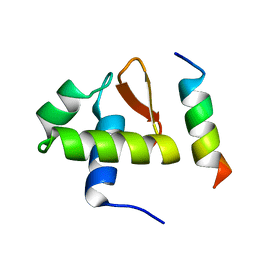 | | SOLUTION STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN RPA32 COMPLEXED WITH UNG2(73-88) | | Descriptor: | REPLICATION PROTEIN A (RPA32) C-TERMINAL DOMAIN, URACIL DNA GLYCOSYLASE (UNG2) | | Authors: | Mer, G, Edwards, A.M, Chazin, W.J. | | Deposit date: | 1999-12-27 | | Release date: | 2000-11-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA.
Cell(Cambridge,Mass.), 103, 2000
|
|
1PMC
 
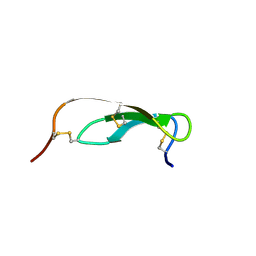 | |
4QO6
 
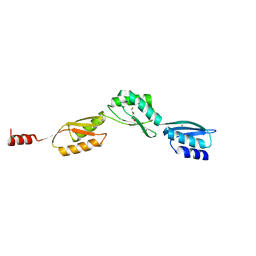 | |
4QQ0
 
 | |
2WKT
 
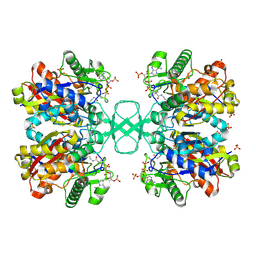 | | BIOSYNTHETIC THIOLASE FROM Z. RAMIGERA. COMPLEX OF THE N316A MUTANT WITH COENZYME A. | | Descriptor: | ACETYL-COA ACETYLTRANSFERASE, CHLORIDE ION, COENZYME A, ... | | Authors: | Merilainen, G, Poikela, V, Kursula, P, Wierenga, R.K. | | Deposit date: | 2009-06-18 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Thiolase Reaction Mechanism: The Importance of Asn316 and His348 for Stabilizing the Enolate Intermediate of the Claisen Condensation.
Biochemistry, 48, 2009
|
|
2WL6
 
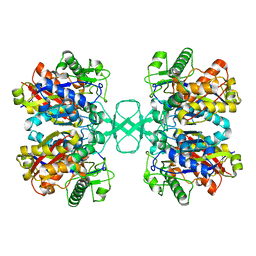 | |
2WL5
 
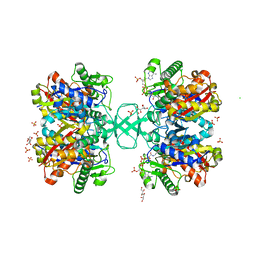 | | BIOSYNTHETIC THIOLASE FROM Z. RAMIGERA. COMPLEX OF THE H348N MUTANT WITH COENZYME A. | | Descriptor: | ACETYL-COA ACETYLTRANSFERASE, CHLORIDE ION, COENZYME A, ... | | Authors: | Merilainen, G, Poikela, V, Kursula, P, Wierenga, R.K. | | Deposit date: | 2009-06-22 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Thiolase Reaction Mechanism: The Importance of Asn316 and His348 for Stabilizing the Enolate Intermediate of the Claisen Condensation.
Biochemistry, 48, 2009
|
|
2WKV
 
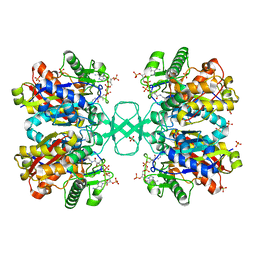 | | BIOSYNTHETIC THIOLASE FROM Z. RAMIGERA. COMPLEX OF THE N316D MUTANT WITH COENZYME A. | | Descriptor: | ACETYL-COA ACETYLTRANSFERASE, COENZYME A, SODIUM ION, ... | | Authors: | Merilainen, G, Poikela, V, Kursula, P, Wierenga, R.K. | | Deposit date: | 2009-06-18 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Thiolase Reaction Mechanism: The Importance of Asn316 and His348 for Stabilizing the Enolate Intermediate of the Claisen Condensation.
Biochemistry, 48, 2009
|
|
2WKU
 
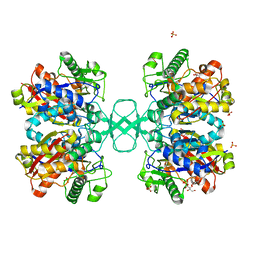 | | BIOSYNTHETIC THIOLASE FROM Z. RAMIGERA. THE N316H MUTANT. | | Descriptor: | ACETYL-COA ACETYLTRANSFERASE, D-mannose, SULFATE ION | | Authors: | Merilainen, G, Poikela, V, Kursula, P, Wierenga, R.K. | | Deposit date: | 2009-06-18 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Thiolase Reaction Mechanism: The Importance of Asn316 and His348 for Stabilizing the Enolate Intermediate of the Claisen Condensation.
Biochemistry, 48, 2009
|
|
2WL4
 
 | | BIOSYNTHETIC THIOLASE FROM Z. RAMIGERA. COMPLEX OF THE H348A MUTANT WITH COENZYME A. | | Descriptor: | ACETYL-COA ACETYLTRANSFERASE, CHLORIDE ION, COENZYME A, ... | | Authors: | Merilainen, G, Poikela, V, Kursula, P, Wierenga, R.K. | | Deposit date: | 2009-06-22 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Thiolase Reaction Mechanism: The Importance of Asn316 and His348 for Stabilizing the Enolate Intermediate of the Claisen Condensation.
Biochemistry, 48, 2009
|
|
2K3Y
 
 | |
2K3X
 
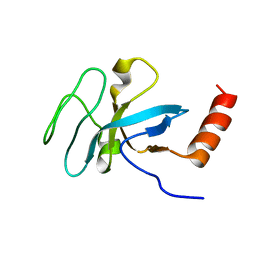 | | Solution structure of EAF3 chromo barrel domain | | Descriptor: | Chromatin modification-related protein EAF3 | | Authors: | Mer, G, Xu, C. | | Deposit date: | 2008-05-19 | | Release date: | 2008-09-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Recognition of Methylated Histone H3K36 by the Eaf3 Subunit of Histone Deacetylase Complex Rpd3S.
Structure, 16, 2008
|
|
2IG0
 
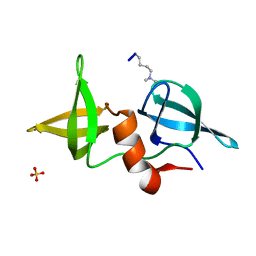 | | Structure of 53BP1/methylated histone peptide complex | | Descriptor: | Dimethylated Histone H4-K20 peptide, SULFATE ION, Tumor suppressor p53-binding protein 1 | | Authors: | Mer, G. | | Deposit date: | 2006-09-22 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Methylation State-Specific Recognition of Histone H4-K20 by 53BP1 and Crb2 in DNA Repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
2VU2
 
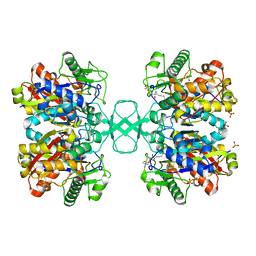 | | Biosynthetic thiolase from Z. ramigera. Complex with S-pantetheine-11- pivalate. | | Descriptor: | (3R)-3-hydroxy-2,2-dimethyl-4-oxo-4-({3-oxo-3-[(2-sulfanylethyl)amino]propyl}amino)butyl 2,2-dimethylpropanoate, ACETYL-COA ACETYLTRANSFERASE, SULFATE ION | | Authors: | Kursula, P, Merilainen, G, Schmitz, W, Wierenga, R.K. | | Deposit date: | 2008-05-19 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Sulfur Atoms of the Substrate Coa and the Catalytic Cysteine are Required for a Productive Mode of Substrate Binding in Bacterial Biosynthetic Thiolase, a Thioester-Dependent Enzyme.
FEBS J., 275, 2008
|
|
2VTZ
 
 | | Biosynthetic thiolase from Z. ramigera. Complex of the C89A mutant with coenzyme A. | | Descriptor: | ACETYL-COA ACETYLTRANSFERASE, COENZYME A, SULFATE ION | | Authors: | Kursula, P, Merilainen, G, Wierenga, R.K. | | Deposit date: | 2008-05-19 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Sulfur Atoms of the Substrate Coa and the Catalytic Cysteine are Required for a Productive Mode of Substrate Binding in Bacterial Biosynthetic Thiolase, a Thioester-Dependent Enzyme.
FEBS J., 275, 2008
|
|
1HV2
 
 | | SOLUTION STRUCTURE OF YEAST ELONGIN C IN COMPLEX WITH A VON HIPPEL-LINDAU PEPTIDE | | Descriptor: | ELONGIN C, VON HIPPEL-LINDAU DISEASE TUMOR SUPPRESSOR | | Authors: | Botuyan, M.V, Mer, G, Yi, G.-S, Koth, C.M, Case, D.A, Edwards, A.M, Chazin, W.J, Arrowsmith, C.H. | | Deposit date: | 2001-01-05 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of yeast elongin C in complex with a von Hippel-Lindau peptide.
J.Mol.Biol., 312, 2001
|
|
4UV9
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Ethyl-Tranylcypromine | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(4aS,10aS)-4a-[(1S,3E)-3-imino-1-phenylpentyl]-7,8-dimethyl-2,4-dioxo-1,3,4,4a,5,10a-hexahydrobenzo[g]pteridin-10(2H)-yl]pentyl dihydrogen diphosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
4UVB
 
 | | LSD1(KDM1A)-CoREST in complex with 1-Methyl-Tranylcypromine (1S,2R) | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-[(1R,3S,3aS,7aS)-1-amino-1,10,11-trimethyl-4,6-dioxo-3-phenyl-2,3,5,6,7,7a-hexahydro-1H-benzo[g]pyrrolo[2,1-e]pteridin-8(4H)-yl]-2,3,4-trihydroxypentyl dihydrogen diphosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
4V2D
 
 | | FLRT2 LRR domain | | Descriptor: | FIBRONECTIN LEUCINE RICH TRANSMEMBRANE PROTEIN 2 | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2015-07-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
3ZRM
 
 | | Identification of 2-(4-pyridyl)thienopyridinones as GSK-3beta inhibitors | | Descriptor: | 7-(4-HYDROXYPHENYL)-2-PYRIDIN-4-YL-5H-THIENO[3,2-C]PYRIDIN-4-ONE, GLYCEROL, GLYCOGEN SYNTHASE KINASE-3 BETA, ... | | Authors: | Gentile, G, Bernasconi, G, Pozzan, A, Merlo, G, Marzorati, P, Bamborough, P, Bax, B, Bridges, A, Brough, C, Carter, P, Cutler, G, Neu, M, Takada, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Identification of 2-(4-Pyridyl)Thienopyridinones as Gsk-3Beta Inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3ZRK
 
 | | Identification of 2-(4-pyridyl)thienopyridinones as GSK-3beta inhibitors | | Descriptor: | 2-(4-PYRIDINYL)FURO[3,2-C]PYRIDIN-4(5H)-ONE, GLYCEROL, GLYCOGEN SYNTHASE KINASE-3 BETA, ... | | Authors: | Gentile, G, Bernasconi, G, Pozzan, A, Merlo, G, Marzorati, P, Bamborough, P, Bax, B, Bridges, A, Brough, C, Carter, P, Cutler, G, Neu, M, Takada, M. | | Deposit date: | 2011-06-16 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Identification of 2-(4-Pyridyl)Thienopyridinones as Gsk-3Beta Inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3ZRL
 
 | | Identification of 2-(4-pyridyl)thienopyridinones as GSK-3beta inhibitors | | Descriptor: | 7-BROMO-2-PYRIDIN-4-YL-5H-THIENO[3,2-C]PYRIDIN-4-ONE, GLYCEROL, GLYCOGEN SYNTHASE KINASE-3 BETA, ... | | Authors: | Gentile, G, Bernasconi, G, Pozzan, A, Merlo, G, Marzorati, P, Bamborough, P, Bax, B, Bridges, A, Brough, C, Carter, P, Cutler, G, Neu, M, Takada, M. | | Deposit date: | 2011-06-17 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Identification of 2-(4-Pyridyl)Thienopyridinones as Gsk-3Beta Inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6EJI
 
 | | Structure of a glycosyltransferase | | Descriptor: | CHLORIDE ION, GLYCEROL, POTASSIUM ION, ... | | Authors: | Ramirez, A.S, Boilevin, J, Mehdipour, A.R, Hummer, G, Darbre, T, Reymond, J.L, Locher, K.P. | | Deposit date: | 2017-09-21 | | Release date: | 2018-02-07 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase.
Nat Commun, 9, 2018
|
|
6EJK
 
 | | Structure of a glycosyltransferase | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-glucopyranose, NerylNeryl pyrophosphate, Uridine-Diphosphate-Methylene-N-acetyl-galactosamine, ... | | Authors: | Ramirez, A.S, Boilevin, J, Mehdipour, A.R, Hummer, G, Darbre, T, Reymond, J.L, Locher, K.P. | | Deposit date: | 2017-09-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase.
Nat Commun, 9, 2018
|
|
6MXY
 
 | |
