6YDP
 
 | |
6YDW
 
 | |
6GB2
 
 | | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. This file contains the 39S ribosomal subunit. | | Descriptor: | 'Mitochondrial ribosomal protein L30, 'Mitochondrial ribosomal protein L55, 'Mitochondrial ribosomal protein L59, ... | | Authors: | Kummer, E, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2018-04-13 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Nature, 560, 2018
|
|
6GAW
 
 | | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. This file contains the complete 55S ribosome. | | Descriptor: | 12S ribosomal RNA, mitochondrial, 16S ribosomal RNA, ... | | Authors: | Kummer, E, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2018-04-13 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Nature, 560, 2018
|
|
6GAZ
 
 | | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. This file contains the 28S ribosomal subunit. | | Descriptor: | 12S ribosomal RNA, mitochondrial, 28S ribosomal protein S18b, ... | | Authors: | Kummer, E, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2018-04-13 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Nature, 560, 2018
|
|
1HUQ
 
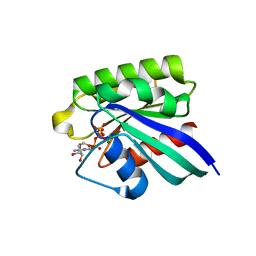 | | 1.8A CRYSTAL STRUCTURE OF THE MONOMERIC GTPASE RAB5C (MOUSE) | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, RAB5C | | Authors: | Merithew, E, Hatherly, S, Dumas, J.J, Lawe, D.C, Heller-Harrison, R, Lambright, D.G. | | Deposit date: | 2001-01-04 | | Release date: | 2001-02-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural plasticity of an invariant hydrophobic triad in the switch regions of Rab GTPases is a determinant of effector recognition.
J.Biol.Chem., 276, 2001
|
|
7NQL
 
 | | 55S mammalian mitochondrial ribosome with ICT1 and P site tRNAMet | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S16, ... | | Authors: | Kummer, E, Schubert, K, Ban, N. | | Deposit date: | 2021-03-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of translation termination, rescue, and recycling in mammalian mitochondria.
Mol.Cell, 81, 2021
|
|
7NSH
 
 | | 39S mammalian mitochondrial large ribosomal subunit with mtRRF (post) and mtEFG2 | | Descriptor: | 16S rRNA, 39S ribosomal protein L48, mitochondrial, ... | | Authors: | Kummer, E, Schubert, K, Ban, N. | | Deposit date: | 2021-03-07 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of translation termination, rescue, and recycling in mammalian mitochondria.
Mol.Cell, 81, 2021
|
|
7NQH
 
 | | 55S mammalian mitochondrial ribosome with mtRF1a and P-site tRNAMet | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S16, ... | | Authors: | Kummer, E, Schubert, K, Ban, N. | | Deposit date: | 2021-03-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of translation termination, rescue, and recycling in mammalian mitochondria.
Mol.Cell, 81, 2021
|
|
7NSI
 
 | | 55S mammalian mitochondrial ribosome with mtRRF (pre) and tRNA(P/E) | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S10, ... | | Authors: | Kummer, E, Schubert, K, Ban, N. | | Deposit date: | 2021-03-07 | | Release date: | 2021-06-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of translation termination, rescue, and recycling in mammalian mitochondria.
Mol.Cell, 81, 2021
|
|
7NSJ
 
 | | 55S mammalian mitochondrial ribosome with tRNA(P/P) and tRNA(E*) | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S16, ... | | Authors: | Kummer, E, Schubert, K, Ban, N. | | Deposit date: | 2021-03-07 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of translation termination, rescue, and recycling in mammalian mitochondria.
Mol.Cell, 81, 2021
|
|
5ONM
 
 | | Crystal Structure of Ectoine Synthase from P. lautus | | Descriptor: | FE (III) ION, L-ectoine synthase, SULFATE ION | | Authors: | Bremer, E. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Illuminating the catalytic core of ectoine synthase through structural and biochemical analysis.
Sci Rep, 9, 2019
|
|
5ONN
 
 | | Crystal Structure of Ectoine Synthase from P. lautus | | Descriptor: | (2~{S})-4-acetamido-2-azanyl-butanoic acid, FE (III) ION, L-ectoine synthase | | Authors: | Bremer, E. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Illuminating the catalytic core of ectoine synthase through structural and biochemical analysis.
Sci Rep, 9, 2019
|
|
5ONO
 
 | | Crystal Structure of Ectoine Synthase from P. lautus | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, FE (III) ION, L-ectoine synthase | | Authors: | Bremer, E. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Illuminating the catalytic core of ectoine synthase through structural and biochemical analysis.
Sci Rep, 9, 2019
|
|
8PK0
 
 | | human mitoribosomal large subunit assembly intermediate 1 with GTPBP10-GTPBP7 | | Descriptor: | 16S rRNA + pre-H68-71 segment, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Kummer, E, Nguyen, T.G, Ritter, C. | | Deposit date: | 2023-06-23 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights into the role of GTPBP10 in the RNA maturation of the mitoribosome.
Nat Commun, 14, 2023
|
|
3RCJ
 
 | | Rapid preparation of triazolyl substituted NH-heterocyclic kinase inhibitors via one-pot Sonogashira coupling TMS-deprotection CuAAC sequence | | Descriptor: | 3-(1-benzyl-1H-1,2,3-triazol-4-yl)-1H-pyrrolo[2,3-b]pyridine, 3-phosphoinositide-dependent protein kinase 1 | | Authors: | Graedler, U, Dorsch, D, Merkul, E, Klukas, F, Sirrenberg, C, Greiner, H.E, Mueller, T.J.J. | | Deposit date: | 2011-03-31 | | Release date: | 2011-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rapid preparation of triazolyl substituted NH-heterocyclic kinase inhibitors via one-pot Sonogashira coupling-TMS-deprotection-CuAAC sequence.
Org.Biomol.Chem., 9, 2011
|
|
3URO
 
 | | Poliovirus receptor CD155 D1D2 | | Descriptor: | Poliovirus receptor | | Authors: | Zhang, P, Mueller, S, Morais, M.C, Bator, C.M, Bowman, V.D, Hafenstein, S, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2011-11-22 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5005 Å) | | Cite: | Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7R6X
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2E12 Fab, S309 Fab, and S304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Monoclonal antibody S2E12 Fab heavy chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7R6W
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2X35 Fab and S309 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Snell, G, Czudnochowski, N, Hernandez, P, Nix, J.C, Croll, T.I, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
4U67
 
 | | Crystal structure of the large ribosomal subunit (50S) of Deinococcus radiodurans containing a three residue insertion in L22 | | Descriptor: | 23s RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A. | | Deposit date: | 2014-07-28 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The Ribosomal Protein uL22 Modulates the Shape of the Protein Exit Tunnel.
Structure, 25, 2017
|
|
7LYC
 
 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A Lys13 and Lys15 in complex with BARD1 (residues 415-777) | | Descriptor: | BRCA1-associated RING domain protein 1, DNA (146-MER), DNA (147-MER), ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, D, Mer, E, Mer, G. | | Deposit date: | 2021-03-06 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation.
Nature, 596, 2021
|
|
4UE3
 
 | | The Mechanism of Hydrogen Activation by NiFe-hydrogenases and the Importance of the active site Arginine | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Evans, R.M, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of Hydrogen Activation by [Nife] Hydrogenases.
Nat.Chem.Biol., 12, 2016
|
|
7LYA
 
 | | Cryo-EM structure of the human nucleosome core particle with linked histone proteins H2A and H2B | | Descriptor: | DNA (146-MER), DNA (147-MER), Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, D, Mer, E, Mer, G. | | Deposit date: | 2021-03-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation.
Nature, 596, 2021
|
|
7LYB
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with BRCA1-BARD1-UbcH5c | | Descriptor: | BRCA1-associated RING domain protein 1, DNA (146-MER), DNA (147-MER), ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, D, Mer, E, Mer, G. | | Deposit date: | 2021-03-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation.
Nature, 596, 2021
|
|
4WFN
 
 | | Crystal structure of the large ribosomal subunit (50S) of Deinococcus radiodurans containing a three residue insertion in L22 in complex with erythromycin | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Wekselman, I, Zimmerman, E, Rozenberg, H, Bashan, A, Yonath, A. | | Deposit date: | 2014-09-16 | | Release date: | 2015-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | The Ribosomal Protein uL22 Modulates the Shape of the Protein Exit Tunnel.
Structure, 25, 2017
|
|
