7QDH
 
 | | SARS-CoV-2 S protein S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7QDG
 
 | | SARS-CoV-2 S protein S:A222V + S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7KVC
 
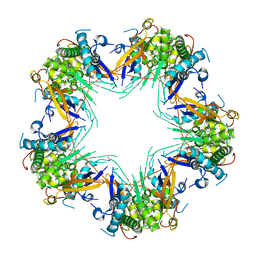 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (decamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
7KVD
 
 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (dodecamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
7R4Q
 
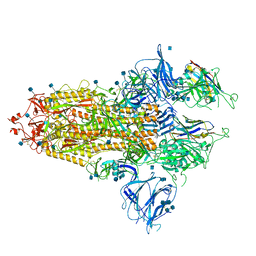 | | The SARS-CoV-2 spike in complex with the 1.29 neutralizing nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camel-derived nanobody 1.29, ... | | Authors: | Casasnovas, J.M, Melero, R, Arranz, R, Fernandez, L.A. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron.
Front Immunol, 13, 2022
|
|
7R4R
 
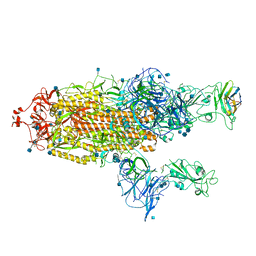 | | The SARS-CoV-2 spike in complex with the 1.10 neutralizing nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camel-derived nanobody 1.10, ... | | Authors: | Casasnovas, J.M, Melero, R, Arranz, R, Fernandez, L.A. | | Deposit date: | 2022-02-09 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron.
Front Immunol, 13, 2022
|
|
7R4I
 
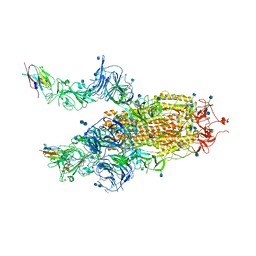 | | The SARS-CoV-2 spike in complex with the 2.15 neutralizing nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Camel-derived nanobody 2.15, ... | | Authors: | Casasnovas, J.M, Melero, R, Arranz, R, Fernandez, L.A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Nanobodies Protecting From Lethal SARS-CoV-2 Infection Target Receptor Binding Epitopes Preserved in Virus Variants Other Than Omicron.
Front Immunol, 13, 2022
|
|
7JSR
 
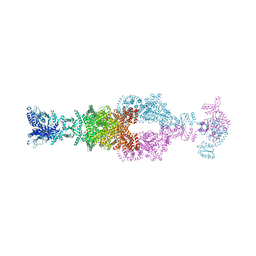 | | Crystal structure of the large glutamate dehydrogenase composed of 180 kDa subunits from Mycobacterium smegmatis | | Descriptor: | NAD-specific glutamate dehydrogenase | | Authors: | Lazaro, M, Melero, R, Huet, C, Lopez-Alonso, J.P, Delgado, S, Dodu, A, Bruch, E.M, Abriata, L.A, Alzari, P.M, Valle, M, Lisa, M.N. | | Deposit date: | 2020-08-15 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (6.27 Å) | | Cite: | 3D architecture and structural flexibility revealed in the subfamily of large glutamate dehydrogenases by a mycobacterial enzyme.
Commun Biol, 4, 2021
|
|
7A1D
 
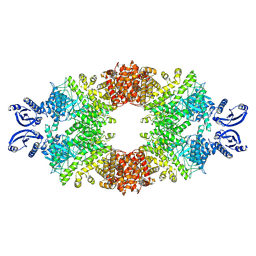 | | Cryo-EM map of the large glutamate dehydrogenase composed of 180 kDa subunits from Mycobacterium smegmatis (open conformation) | | Descriptor: | NAD-specific glutamate dehydrogenase | | Authors: | Lazaro, M, Melero, R, Huet, C, Lopez-Alonso, J.P, Delgado, S, Dodu, A, Bruch, E.M, Abriata, L.A, Alzari, P.M, Valle, M, Lisa, M.N. | | Deposit date: | 2020-08-12 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | 3D architecture and structural flexibility revealed in the subfamily of large glutamate dehydrogenases by a mycobacterial enzyme.
Commun Biol, 4, 2021
|
|
6ZP7
 
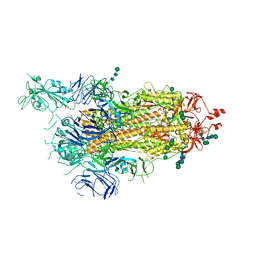 | | SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up open conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
6ZOW
 
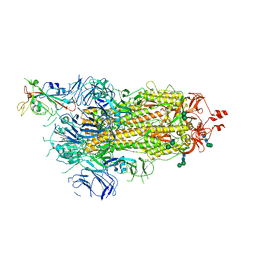 | | SARS-CoV-2 spike in prefusion state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
6ZP5
 
 | | SARS-CoV-2 spike in prefusion state (flexibility analysis, 1-up closed conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Martinez, M, Marabini, R, Carazo, J.M. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Continuous flexibility analysis of SARS-CoV-2 spike prefusion structures.
Iucrj, 7, 2020
|
|
8P9Y
 
 | | SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | Deposit date: | 2023-06-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
8P99
 
 | | SARS-CoV-2 S-protein:D614G mutant in 1-up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1,Spike glycoprotein | | Authors: | Adhav, A, Forcada-Nadal, A, Marco-Marin, C, Lopez-Redondo, M.L, Llacer, J.L. | | Deposit date: | 2023-06-05 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | C-2 Thiophenyl Tryptophan Trimers Inhibit Cellular Entry of SARS-CoV-2 through Interaction with the Viral Spike (S) Protein.
J.Med.Chem., 66, 2023
|
|
8RBY
 
 | |
8RJ7
 
 | |
