8J47
 
 | | CryoEM Structure of 40-Residue Arctic (E22G) Beta-Amyloid Fibril Derived by Co-Analysis with Solid-State NMR | E22G Abeta40 | | Descriptor: | E22G Amyloid-beta | | Authors: | Tehrani, M.J, Matsuda, I, Yamagata, A, Matsunaga, T, Sato, M, Toyooka, K, Shirouzu, M, Ishii, Y, Kodama, Y, McElheny, D, Kobayashi, N. | | Deposit date: | 2023-04-19 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | E22G A beta 40 fibril structure and kinetics illuminate how A beta 40 rather than A beta 42 triggers familial Alzheimer's.
Nat Commun, 15, 2024
|
|
1EHL
 
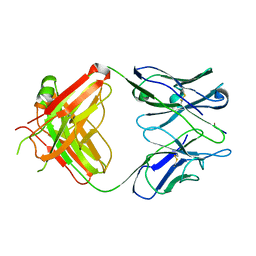 | | 64M-2 ANTIBODY FAB COMPLEXED WITH D(5HT)(6-4)T | | Descriptor: | 5'-(D(5HT)P*(6-4)T)-3', ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (HEAVY CHAIN), ANTI-(6-4) PHOTOPRODUCT ANTIBODY 64M-2 FAB (LIGHT CHAIN) | | Authors: | Yokoyama, H, Mizutani, R, Satow, Y, Komatsu, Y, Ohtsuka, E, Nikaido, O. | | Deposit date: | 2000-02-21 | | Release date: | 2001-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the 64M-2 antibody Fab fragment in complex with a DNA dT(6-4)T photoproduct formed by ultraviolet radiation.
J.Mol.Biol., 299, 2000
|
|
3QKZ
 
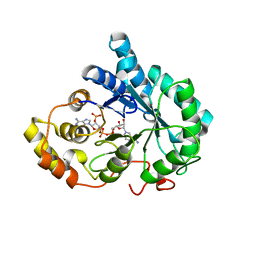 | | Crystal structure of mutant His269Arg AKR1B14 | | Descriptor: | Aldo-keto reductase family 1, member B7, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaram, K, El-Kabbani, O. | | Deposit date: | 2011-02-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of the His269Arg mutant of the rat aldose reductase-like protein AKR1B14 complexed with NADPH.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3O3R
 
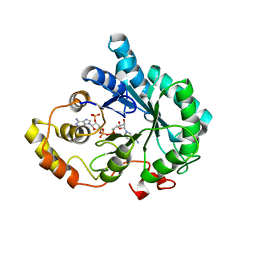 | | Crystal Structure of AKR1B14 in complex with NADP | | Descriptor: | Aldo-keto reductase family 1, member B7, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaram, K, Dhagat, U, El-Kabbani, O. | | Deposit date: | 2010-07-26 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of rat aldose reductase-like protein AKR1B14 holoenzyme: Probing the role of His269 in coenzyme binding by site-directed mutagenesis
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3NTY
 
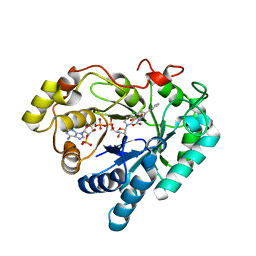 | | Crystal structure of AKR1C1 in complex with NADP and 5-Phenyl,3-chlorosalicylic acid | | Descriptor: | 5-chloro-4-hydroxybiphenyl-3-carboxylic acid, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Dhagat, U, El-Kabbani, O. | | Deposit date: | 2010-07-06 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Probing the inhibitor selectivity pocket of human 20 alpha-hydroxysteroid dehydrogenase (AKR1C1) with X-ray crystallography and site-directed mutagenesis
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7C7G
 
 | |
1GZB
 
 | | PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PEROXIDASE, ... | | Authors: | Fukuyama, K, Kunishima, N, Amada, F. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pentacoordination of the heme iron of Arthromyces ramosus peroxidase shown by a 1.8 A resolution crystallographic study at pH 4.5.
FEBS Lett., 378, 1996
|
|
7C7F
 
 | | Crystal structures of AKR1C3 binary complex with NADP+ | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Aldo-keto reductase family 1 member C3, ... | | Authors: | Irie, K, Toyooka, N, Endo, S. | | Deposit date: | 2020-05-25 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of Novel AKR1C3 Inhibitors as New Potential Treatment for Castration-Resistant Prostate Cancer.
J.Med.Chem., 63, 2020
|
|
7C7H
 
 | |
1HSS
 
 | | 0.19 ALPHA-AMYLASE INHIBITOR FROM WHEAT | | Descriptor: | 0.19 ALPHA-AMYLASE INHIBITOR | | Authors: | Oda, Y, Fukuyama, K. | | Deposit date: | 1997-07-01 | | Release date: | 1998-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Tertiary and quaternary structures of 0.19 alpha-amylase inhibitor from wheat kernel determined by X-ray analysis at 2.06 A resolution.
Biochemistry, 36, 1997
|
|
1C8I
 
 | | BINDING MODE OF HYDROXYLAMINE TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HYDROXYAMINE, ... | | Authors: | Wariishi, H, Nonaka, D, Johjima, T, Nakamura, N, Naruta, Y, Kubo, K, Fukuyama, K. | | Deposit date: | 2000-05-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct binding of hydroxylamine to the heme iron of Arthromyces ramosus peroxidase. Substrate analogue that inhibits compound I formation in a competetive manner.
J.Biol.Chem., 275, 2000
|
|
1CK6
 
 | | BINDING MODE OF SALICYLHYDROXAMIC ACID TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PROTEIN (PEROXIDASE), ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1999-04-28 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding of salicylhydroxamic acid and several aromatic donor molecules to Arthromyces ramosus peroxidase, investigated by X-ray crystallography, optical difference spectroscopy, NMR relaxation, molecular dynamics, and kinetics.
Biochemistry, 38, 1999
|
|
2O48
 
 | | Crystal structure of Mammalian Dimeric Dihydrodiol Dehydrogenase | | Descriptor: | BETA-MERCAPTOETHANOL, Dimeric dihydrodiol dehydrogenase, P-HYDROXYACETOPHENONE, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2006-12-04 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structures of dimeric dihydrodiol dehydrogenase apoenzyme and inhibitor complex: probing the subunit interface with site-directed mutagenesis.
Proteins, 70, 2008
|
|
2O4U
 
 | | Crystal structure of Mammalian Dimeric Dihydrodiol Dehydrogenase | | Descriptor: | BETA-MERCAPTOETHANOL, Dimeric dihydrodiol dehydrogenase, GLYCEROL, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2006-12-04 | | Release date: | 2007-07-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of dimeric dihydrodiol dehydrogenase apoenzyme and inhibitor complex: probing the subunit interface with site-directed mutagenesis.
Proteins, 70, 2008
|
|
1GZA
 
 | | PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1996-11-13 | | Release date: | 1997-03-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Binding of iodide to Arthromyces ramosus peroxidase investigated with X-ray crystallographic analysis, 1H and 127I NMR spectroscopy, and steady-state kinetics.
J.Biol.Chem., 272, 1997
|
|
1HSR
 
 | | BINDING MODE OF BENZHYDROXAMIC ACID TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BENZHYDROXAMIC ACID, CALCIUM ION, ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1997-07-01 | | Release date: | 1998-07-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding mode of benzhydroxamic acid to Arthromyces ramosus peroxidase shown by X-ray crystallographic analysis of the complex at 1.6 A resolution.
FEBS Lett., 412, 1997
|
|
1KEG
 
 | | Antibody 64M-2 Fab complexed with dTT(6-4)TT | | Descriptor: | 5'-D(*TP*(64T)P*TP*T)-3', Anti-(6-4) photoproduct antibody 64M-2 Fab (heavy chain), Anti-(6-4) photoproduct antibody 64M-2 Fab (light chain), ... | | Authors: | Yokoyama, H, Mizutani, R, Satow, Y, Sato, K, Komatsu, Y, Ohtsuka, E, Nikaido, O. | | Deposit date: | 2001-11-15 | | Release date: | 2002-11-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the DNA (6-4) photoproduct dTT(6-4)TT in complex with the 64M-2 antibody Fab fragment implies increased antibody-binding affinity by the flanking nucleotides.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2D1Y
 
 | |
