3TG6
 
 | | Crystal Structure of Influenza A Virus nucleoprotein with Ligand | | Descriptor: | Nucleocapsid protein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl][3-(2-chloropyridin-3-yl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Pearce, B.C, Lewis, H.A, McDonnell, P.A, Steinbacher, S, Kiefersauer, R, Mortl, M, Maskos, K, Edavettal, S, Baldwin, E.T, Langley, D.R. | | Deposit date: | 2011-08-17 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biophysical and Structural Characterization of a Novel Class of Influenza Virus Inhibitors
To be Published
|
|
1BUV
 
 | | CRYSTAL STRUCTURE OF THE MT1-MMP-TIMP-2 COMPLEX | | Descriptor: | CALCIUM ION, PROTEIN (MEMBRANE-TYPE MATRIX METALLOPROTEINASE (CDMT1-MMP)), PROTEIN (METALLOPROTEINASE INHIBITOR (TIMP-2)), ... | | Authors: | Fernandez-Catalan, C, Bode, W, Huber, R, Turk, D, Calvete, J.J, Lichte, A, Tschesche, H, Maskos, K. | | Deposit date: | 1998-09-07 | | Release date: | 1999-09-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor.
EMBO J., 17, 1998
|
|
1BQQ
 
 | | CRYSTAL STRUCTURE OF THE MT1-MMP--TIMP-2 COMPLEX | | Descriptor: | CALCIUM ION, MEMBRANE-TYPE MATRIX METALLOPROTEINASE, METALLOPROTEINASE INHIBITOR 2, ... | | Authors: | Fernandez-Catalan, C, Bode, W, Huber, R, Turk, D, Calvete, J.J, Lichte, A, Tschesche, H, Maskos, K. | | Deposit date: | 1998-08-18 | | Release date: | 1999-08-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor.
EMBO J., 17, 1998
|
|
3V3K
 
 | |
4OD9
 
 | | Structure of Cathepsin D with inhibitor N-(3,4-dimethoxybenzyl)-Nalpha-{N-[(3,4-dimethoxyphenyl)acetyl]carbamimidoyl}-D-phenylalaninamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Graedler, U, Czodrowski, P, Tsaklakidis, C, Klein, M, Maskos, K, Leuthner, B. | | Deposit date: | 2014-01-10 | | Release date: | 2014-08-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based optimization of non-peptidic Cathepsin D inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4OC6
 
 | | Structure of Cathepsin D with inhibitor 2-bromo-N-[(2S,3S)-4-{[2-(2,4-dichlorophenyl)ethyl][3-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)propanoyl]amino}-3-hydroxy-1-(3-phenoxyphenyl)butan-2-yl]-4,5-dimethoxybenzamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-bromo-N-[(2S,3S)-4-{[2-(2,4-dichlorophenyl)ethyl][3-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)propanoyl]amino}-3-hydroxy-1-(3-phenoxyphenyl)butan-2-yl]-4,5-dimethoxybenzamide, Cathepsin D heavy chain, ... | | Authors: | Graedler, U, Czodrowski, P, Tsaklakidis, C, Klein, M, Maskos, K, Leuthner, B. | | Deposit date: | 2014-01-08 | | Release date: | 2014-08-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure-based optimization of non-peptidic Cathepsin D inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4OBZ
 
 | | Structure of Cathepsin D with inhibitor 2-(3,4-dimethoxyphenyl)-N-[N-(4-methylbenzyl)carbamimidoyl]acetamide | | Descriptor: | 2-(3,4-dimethoxyphenyl)-N-[N-(4-methylbenzyl)carbamimidoyl]acetamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Graedler, U, Czodrowski, P, Tsaklakidis, C, Klein, M, Maskos, K, Leuthner, B. | | Deposit date: | 2014-01-08 | | Release date: | 2014-08-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based optimization of non-peptidic Cathepsin D inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1JK3
 
 | | Crystal structure of human MMP-12 (Macrophage Elastase) at true atomic resolution | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, MACROPHAGE METALLOELASTASE, ... | | Authors: | Lang, R, Kocourek, A, Braun, M, Tschesche, H, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2001-07-11 | | Release date: | 2001-09-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Substrate specificity determinants of human macrophage elastase (MMP-12) based on the 1.1 A crystal structure.
J.Mol.Biol., 312, 2001
|
|
1LFW
 
 | | Crystal structure of pepV | | Descriptor: | 3-[(1-AMINO-2-CARBOXY-ETHYL)-HYDROXY-PHOSPHINOYL]-2-METHYL-PROPIONIC ACID, ZINC ION, pepV | | Authors: | Jozic, D, Bourenkow, G, Bartunik, H, Scholze, H, Dive, V, Henrich, B, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2002-04-12 | | Release date: | 2002-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Dinuclear Zinc Aminopeptidase PepV from Lactobacillus delbrueckii Unravels Its Preference for Dipeptides
Structure, 10, 2002
|
|
4G6L
 
 | | Crystal structure of human CDK8/CYCC in the DMG-in conformation | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, FORMIC ACID | | Authors: | Schneider, E.V, Blaesse, M, Huber, R, Maskos, K. | | Deposit date: | 2012-07-19 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3HGT
 
 | |
3HGQ
 
 | |
1Y79
 
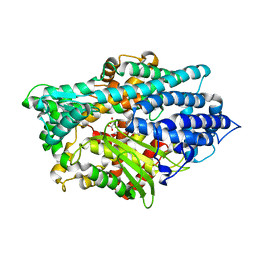 | | Crystal Structure of the E.coli Dipeptidyl Carboxypeptidase Dcp in Complex with a Peptidic Inhibitor | | Descriptor: | ASPARTIC ACID, GLYCINE, LYSINE, ... | | Authors: | Comellas-Bigler, M, Lang, R, Bode, W, Maskos, K. | | Deposit date: | 2004-12-08 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the E.coli Dipeptidyl Carboxypeptidase Dcp: Further Indication of a Ligand-dependant Hinge Movement Mechanism
J.Mol.Biol., 349, 2005
|
|
1T1I
 
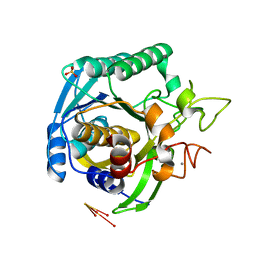 | | High Resolution Crystal Structure of Mutant W129A of Kumamolisin, a Sedolisin Type Proteinase (previously called Kumamolysin or KSCP) | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin | | Authors: | Comellas-Bigler, M, Maskos, K, Huber, R, Oyama, H, Oda, K, Bode, W. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 1.2 a crystal structure of the serine carboxyl proteinase pro-kumamolisin: structure of an intact pro-subtilase
Structure, 12, 2004
|
|
1T1G
 
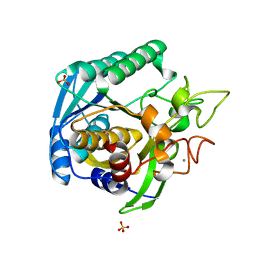 | | High Resolution Crystal Structure of Mutant E23A of Kumamolisin, a sedolisin type proteinase (previously called Kumamolysin or KSCP) | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin | | Authors: | Comellas-Bigler, M, Maskos, K, Huber, R, Oyama, H, Oda, K, Bode, W. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | 1.2 a crystal structure of the serine carboxyl proteinase pro-kumamolisin: structure of an intact pro-subtilase
Structure, 12, 2004
|
|
1T1E
 
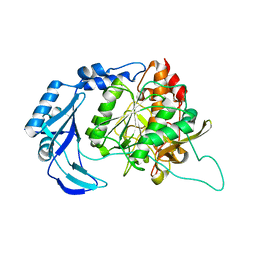 | | High Resolution Crystal Structure of the Intact Pro-Kumamolisin, a Sedolisin Type Proteinase (previously called Kumamolysin or KSCP) | | Descriptor: | CALCIUM ION, kumamolisin | | Authors: | Comellas-Bigler, M, Maskos, K, Huber, R, Oyama, H, Oda, K, Bode, W. | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | 1.2 a crystal structure of the serine carboxyl proteinase pro-kumamolisin: structure of an intact pro-subtilase
Structure, 12, 2004
|
|
2NSM
 
 | | Crystal structure of the human carboxypeptidase N (Kininase I) catalytic domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carboxypeptidase N catalytic chain, SULFATE ION | | Authors: | Keil, C, Maskos, K, Than, M, Hoopes, J.T, Huber, R, Tan, F, Deddish, P.A, Erdoes, E.G, Skidgel, R.A, Bode, W. | | Deposit date: | 2006-11-05 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the human carboxypeptidase N (kininase I) catalytic domain
J.Mol.Biol., 366, 2007
|
|
2OVX
 
 | | MMP-9 active site mutant with barbiturate inhibitor | | Descriptor: | 5-(4-PHENOXYPHENYL)-5-(4-PYRIMIDIN-2-YLPIPERAZIN-1-YL)PYRIMIDINE-2,4,6(2H,3H)-TRIONE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tochowicz, A, Bode, W, Maskos, K, Goettig, P. | | Deposit date: | 2007-02-15 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of MMP-9 Complexes with Five Inhibitors: Contribution of the Flexible Arg424 Side-chain to Selectivity.
J.Mol.Biol., 371, 2007
|
|
2OW0
 
 | | MMP-9 active site mutant with iodine-labeled carboxylate inhibitor | | Descriptor: | CALCIUM ION, CHLORIDE ION, Matrix metalloproteinase-9 (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB), ... | | Authors: | Tochowicz, A, Bode, W, Maskos, K, Goettig, P. | | Deposit date: | 2007-02-15 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of MMP-9 Complexes with Five Inhibitors: Contribution of the Flexible Arg424 Side-chain to Selectivity.
J.Mol.Biol., 371, 2007
|
|
2OW2
 
 | | MMP-9 active site mutant with difluoro butanoic acid inhibitor | | Descriptor: | (3R)-4,4-DIFLUORO-3-[(4-METHOXYPHENYL)SULFONYL]BUTANOIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tochowicz, A, Bode, W, Maskos, K, Goettig, P. | | Deposit date: | 2007-02-15 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of MMP-9 Complexes with Five Inhibitors: Contribution of the Flexible Arg424 Side-chain to Selectivity.
J.Mol.Biol., 371, 2007
|
|
2OW1
 
 | | MMP-9 active site mutant with trifluoromethyl hydroxamate inhibitor | | Descriptor: | (2R)-2-AMINO-3,3,3-TRIFLUORO-N-HYDROXY-2-{[(4-PHENOXYPHENYL)SULFONYL]METHYL}PROPANAMIDE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tochowicz, A, Bode, W, Maskos, K, Goettig, P. | | Deposit date: | 2007-02-15 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of MMP-9 Complexes with Five Inhibitors: Contribution of the Flexible Arg424 Side-chain to Selectivity.
J.Mol.Biol., 371, 2007
|
|
2OVZ
 
 | | MMP-9 active site mutant with phosphinate inhibitor | | Descriptor: | CALCIUM ION, CHLORIDE ION, Matrix metalloproteinase-9 (EC 3.4.24.35) (MMP-9) (92 kDa type IV collagenase) (92 kDa gelatinase) (Gelatinase B) (GELB), ... | | Authors: | Tochowicz, A, Bode, W, Maskos, K, Goettig, P. | | Deposit date: | 2007-02-15 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of MMP-9 Complexes with Five Inhibitors: Contribution of the Flexible Arg424 Side-chain to Selectivity.
J.Mol.Biol., 371, 2007
|
|
2QZW
 
 | |
2QZX
 
 | | Secreted aspartic proteinase (Sap) 5 from Candida albicans | | Descriptor: | Candidapepsin-5, Pepstatin | | Authors: | Lee, J.H, Ruge, E, Borelli, C, Maskos, K, Huber, R. | | Deposit date: | 2007-08-17 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of Sap1 and Sap5: Structural comparison of the secreted aspartic proteinases from Candida albicans.
Proteins, 72, 2008
|
|
1POJ
 
 | | Isoaspartyl Dipeptidase with bound inhibitor | | Descriptor: | 2-{[[(1S)-1-AMINO-2-CARBOXYETHYL](DIHYDROXY)PHOSPHORANYL]METHYL}-4-METHYLPENTANOIC ACID, Isoaspartyl dipeptidase, ZINC ION | | Authors: | Jozic, D, Kaiser, J.T, Huber, R, Bode, W, Maskos, K. | | Deposit date: | 2003-06-15 | | Release date: | 2004-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray structure of isoaspartyl dipeptidase from E.coli: a dinuclear zinc peptidase evolved from amidohydrolases.
J.Mol.Biol., 332, 2003
|
|
