6FO1
 
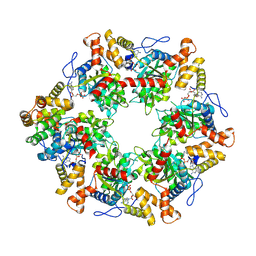 | | Human R2TP subcomplex containing 1 RUVBL1-RUVBL2 hexamer bound to 1 RBD domain from RPAP3. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RNA polymerase II-associated protein 3, RuvB-like 1, ... | | Authors: | Martino, F, Munoz-Hernandez, H, Rodriguez, C.F, Pearl, L.H, Llorca, O. | | Deposit date: | 2018-02-05 | | Release date: | 2018-04-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | RPAP3 provides a flexible scaffold for coupling HSP90 to the human R2TP co-chaperone complex.
Nat Commun, 9, 2018
|
|
6RAZ
 
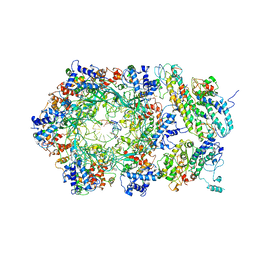 | | D. melanogaster CMG-DNA, State 2B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AT18545p, ... | | Authors: | Eickhoff, P, Martino, F, Costa, A. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-18 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.46 Å) | | Cite: | Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome.
Cell Rep, 28, 2019
|
|
6RAW
 
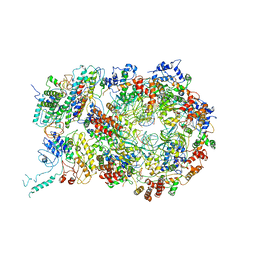 | | D. melanogaster CMG-DNA, State 1A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AT18545p, ... | | Authors: | Eickhoff, P, Martino, F, Costa, A. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome.
Cell Rep, 28, 2019
|
|
6RAX
 
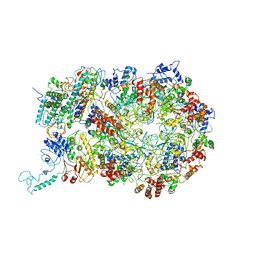 | | D. melanogaster CMG-DNA, State 1B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AT18545p, ... | | Authors: | Eickhoff, P, Martino, F, Costa, A. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome.
Cell Rep, 28, 2019
|
|
6RAY
 
 | | D. melanogaster CMG-DNA, State 2A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, AT18545p, ... | | Authors: | Eickhoff, P, Martino, F, Costa, A. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Molecular Basis for ATP-Hydrolysis-Driven DNA Translocation by the CMG Helicase of the Eukaryotic Replisome.
Cell Rep, 28, 2019
|
|
7P30
 
 | | 3.0 A resolution structure of a DNA-loaded MCM double hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (53-MER), ... | | Authors: | Greiwe, J.F, Miller, T.C.R, Martino, F, Costa, A. | | Deposit date: | 2021-07-06 | | Release date: | 2022-02-02 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mechanism for the selective phosphorylation of DNA-loaded MCM double hexamers by the Dbf4-dependent kinase.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P5Z
 
 | | Structure of a DNA-loaded MCM double hexamer engaged with the Dbf4-dependent kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 7, ... | | Authors: | Greiwe, J.F, Miller, T.C.R, Martino, F, Costa, A. | | Deposit date: | 2021-07-15 | | Release date: | 2022-02-02 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mechanism for the selective phosphorylation of DNA-loaded MCM double hexamers by the Dbf4-dependent kinase.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4LD9
 
 | | Crystal structure of the N-terminally acetylated BAH domain of Sir3 bound to the nucleosome core particle | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3.2, ... | | Authors: | Arnaudo, N, Fernandez, I.S, McLaughlin, S.H, Peak-Chew, S.Y, Rhodes, D, Martino, F. | | Deposit date: | 2013-06-24 | | Release date: | 2013-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.306 Å) | | Cite: | The N-terminal acetylation of Sir3 stabilizes its binding to the nucleosome core particle.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2MWI
 
 | | The structure of the carboxy-terminal domain of DNTTIP1 | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1 | | Authors: | Schwabe, J.W.R, Muskett, F.W, Itoh, T. | | Deposit date: | 2014-11-11 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of a cell cycle associated HDAC1/2 complex reveals the structural basis for complex assembly and nucleosome targeting.
Nucleic Acids Res., 43, 2015
|
|
4D6K
 
 | | Structure of DNTTIP1 dimerisation domain. | | Descriptor: | DEOXYNUCLEOTIDYLTRANSFERASE TERMINAL-INTERACTING PROTEIN 1 | | Authors: | Itoh, T, Fairall, L, Schwabe, J.W.R. | | Deposit date: | 2014-11-11 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of a Cell Cycle Associated Hdac1/2 Complex Reveals the Structural Basis for Complex Assembly and Nucleosome Targeting.
Nucleic Acids Res., 43, 2015
|
|
6Z2K
 
 | | The structure of the tetrameric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
6Z2J
 
 | | The structure of the dimeric HDAC1/MIDEAS/DNTTIP1 MiDAC deacetylase complex | | Descriptor: | Deoxynucleotidyltransferase terminal-interacting protein 1, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Fairall, L, Saleh, A, Ragan, T.J, Millard, C.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-05-16 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The MiDAC histone deacetylase complex is essential for embryonic development and has a unique multivalent structure.
Nat Commun, 11, 2020
|
|
4KXF
 
 | |
