1DY9
 
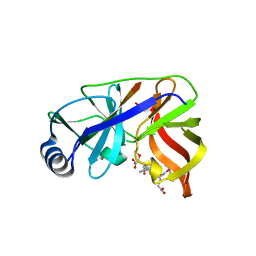 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor I) | | 分子名称: | N-(tert-butoxycarbonyl)-L-alpha-glutamyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70), ... | | 著者 | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | 登録日 | 2000-01-31 | | 公開日 | 2001-01-28 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
1DXP
 
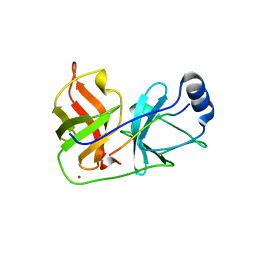 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (apo structure) | | 分子名称: | GLYCEROL, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70), ... | | 著者 | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | 登録日 | 2000-01-13 | | 公開日 | 2001-01-12 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
8XXN
 
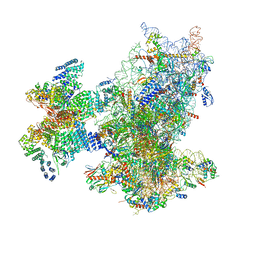 | | Cryo-EM structure of the human 43S ribosome with PDCD4 | | 分子名称: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | 登録日 | 2024-01-18 | | 公開日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 2024
|
|
4UIB
 
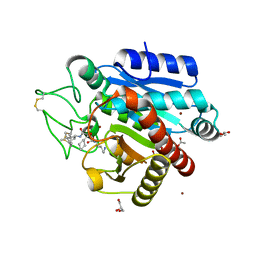 | | Crystal structure of 3p in complex with tafCPB | | 分子名称: | (2S)-6-AMINO-2-[[(1R)-1-(CYCLOHEXYLMETHYL)-2-OXO-2-[[(2S)-1,7,7-TRIMETHYLNORBORNAN-2-YL]AMINO]ETHYL]ARBAMOYLAMINO]HEXANOIC ACID, ACETATE ION, CARBOXYPEPTIDASE B, ... | | 著者 | Halland, N, Broenstrup, M, Czech, J, Czechtizky, W, Evers, A, Follmann, M, Kohlmann, M, Schiell, M, Kurz, M, Schreuder, H.A, Kallus, C. | | 登録日 | 2015-03-27 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Novel Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (Tafia) from Natural Product Anabaenopeptin.
J.Med.Chem., 58, 2015
|
|
4UIA
 
 | | Crystal structure of 3a in complex with tafCPB | | 分子名称: | (2S)-6-azanyl-2-[[(2R)-3-cyclohexyl-1-(3-methylbutylamino)-1-oxidanylidene-propan-2-yl]carbamoylamino]hexanoic acid, CARBOXYPEPTIDASE B, ZINC ION | | 著者 | Halland, N, Broenstrup, M, Czech, J, Czechtizky, W, Evers, A, Follmann, M, Kohlmann, M, Schiell, M, Kurz, M, Schreuder, H.A, Kallus, C. | | 登録日 | 2015-03-27 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Novel Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (Tafia) from Natural Product Anabaenopeptin.
J.Med.Chem., 58, 2015
|
|
8XXL
 
 | | Cryo-EM structure of the human 40S ribosome with PDCD4 | | 分子名称: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | 登録日 | 2024-01-18 | | 公開日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 2024
|
|
8XXM
 
 | | Cryo-EM structure of the human 40S ribosome with PDCD4 and eIF3G | | 分子名称: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | 登録日 | 2024-01-18 | | 公開日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 2024
|
|
1EE8
 
 | | CRYSTAL STRUCTURE OF MUTM (FPG) PROTEIN FROM THERMUS THERMOPHILUS HB8 | | 分子名称: | MUTM (FPG) PROTEIN, ZINC ION | | 著者 | Sugahara, M, Mikawa, T, Kumasaka, T, Yamamoto, M, Kato, R, Fukuyama, K, Inoue, Y, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2000-01-31 | | 公開日 | 2001-01-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8.
EMBO J., 19, 2000
|
|
1EZ0
 
 | | CRYSTAL STRUCTURE OF THE NADP+ DEPENDENT ALDEHYDE DEHYDROGENASE FROM VIBRIO HARVEYI. | | 分子名称: | ALDEHYDE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Ahvazi, B, Coulombe, R, Delarge, M, Vedadi, M, Zhang, L, Meighen, E, Vrielink, A. | | 登録日 | 2000-05-09 | | 公開日 | 2000-05-24 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the NADP+-dependent aldehyde dehydrogenase from Vibrio harveyi: structural implications for cofactor specificity and affinity.
Biochem.J., 349, 2000
|
|
1E94
 
 | | HslV-HslU from E.coli | | 分子名称: | HEAT SHOCK PROTEIN HSLU, HEAT SHOCK PROTEIN HSLV, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Song, H.K, Hartmann, C, Ravishankar, R, Bochtler, M. | | 登録日 | 2000-10-07 | | 公開日 | 2000-11-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mutational Studies on Hslu and its Docking Mode with Hslv
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4USO
 
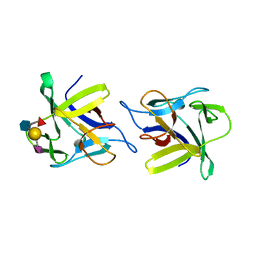 | | X-ray structure of the CCL2 lectin in complex with sialyl lewis X | | 分子名称: | CCL2 LECTIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Bleuler-Martinez, S, Varrot, A, Schubert, M, Stutz, M, Sieber, R, Hengartner, M, Aebi, M, Kunzler, M. | | 登録日 | 2014-07-11 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes.
Glycobiology, 27, 2017
|
|
1DXL
 
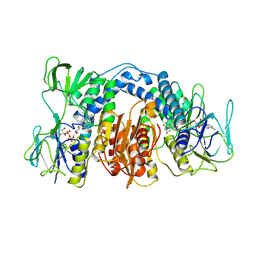 | | Dihydrolipoamide dehydrogenase of glycine decarboxylase from Pisum Sativum | | 分子名称: | DIHYDROLIPOAMIDE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Faure, M, Cohen-Addad, C, Bourguignon, J, Macherel, D, Neuburger, M, Douce, R. | | 登録日 | 2000-01-10 | | 公開日 | 2000-07-20 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Interaction between the Lipoamide-Containing H-Protein and the Lipoamide Dehydrogenase (L-Protein) of the Glycine Decarboxylase Multienzyme System. 2. Crystal Structure of H- and L-Proteins
Eur.J.Biochem., 267, 2000
|
|
1EGN
 
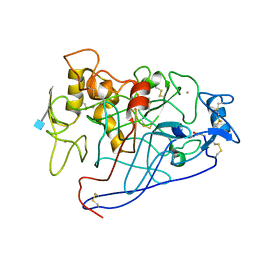 | | CELLOBIOHYDROLASE CEL7A (E223S, A224H, L225V, T226A, D262G) MUTANT | | 分子名称: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE CEL7A, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION | | 著者 | Stahlberg, J, Harris, M, Jones, T.A. | | 登録日 | 2000-02-16 | | 公開日 | 2001-05-16 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Engineering of a glycosidase Family 7 cellobiohydrolase to more alkaline pH optimum: the pH behaviour of Trichoderma reesei Cel7A and its E223S/ A224H/L225V/T226A/D262G mutant.
Biochem.J., 356, 2001
|
|
1EHY
 
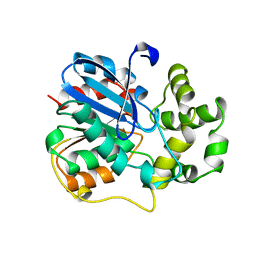 | | X-ray structure of the epoxide hydrolase from agrobacterium radiobacter ad1 | | 分子名称: | POTASSIUM ION, PROTEIN (SOLUBLE EPOXIDE HYDROLASE) | | 著者 | Nardini, M, Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Rink, R, Janssen, D.B, Dijkstra, B.W. | | 登録日 | 1998-10-17 | | 公開日 | 1999-10-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The x-ray structure of epoxide hydrolase from Agrobacterium radiobacter AD1. An enzyme to detoxify harmful epoxides.
J.Biol.Chem., 274, 1999
|
|
1EHJ
 
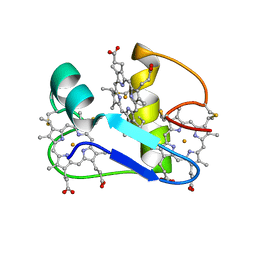 | | A PROTON-NMR INVESTIGATION OF THE FULLY REDUCED CYTOCHROME C7 FROM DESULFUROMONAS ACETOXIDANS | | 分子名称: | CYTOCHROME C7, HEME C | | 著者 | Assfalg, M, Banci, L, Bertini, I, Bruschi, M, Giudici-Orticoni, M.T. | | 登録日 | 2000-02-21 | | 公開日 | 2000-05-10 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A proton-NMR investigation of the fully reduced cytochrome c7 from Desulfuromonas acetoxidans. Comparison between the reduced and the oxidized forms.
Eur.J.Biochem., 266, 1999
|
|
4U01
 
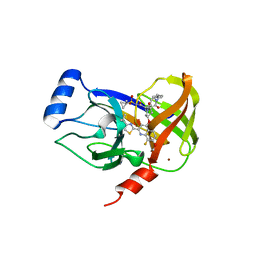 | | HCV NS3/4A serine protease in complex with 6570 | | 分子名称: | (2S,3aS,10Z,11aS,12aR)-2-({8-fluoro-7-methoxy-2-[4-(propan-2-yl)-1,3-thiazol-2-yl]quinolin-4-yl}oxy)-5-methyl-N-[(1-methylcyclopropyl)sulfonyl]-4,14-dioxo-1,2,3,3a,4,5,6,7,8,9,11a,12,13,14-tetradecahydro-12aH-cyclopropa[m]pyrrolo[1,2-c][1,3,6]triazacyclotetradecine-12a-carboxamide, CHLORIDE ION, NS4A protein, ... | | 著者 | Parsy, C.C, Alexandre, F.-R, Brandt, G, Caillet, C, Chaves, D, Derock, M, Gloux, D, Griffon, Y, Lallos, L.B, Leroy, F, Liuzzi, M, Loi, A.-G, Mayes, B, Moulat, L, Moussa, A, Chiara, M, Roques, V, Rosinovsky, E, Seifer, M, Stewart, A, Wang, J, Standring, D, Surleraux, D. | | 登録日 | 2014-07-11 | | 公開日 | 2015-07-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Discovery and structural diversity of the hepatitis C virus NS3/4A serine protease inhibitor series leading to clinical candidate IDX320.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1DTT
 
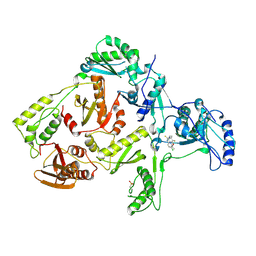 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 (PETT130A94) | | 分子名称: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | 著者 | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | 登録日 | 2000-01-13 | | 公開日 | 2000-04-02 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1DTQ
 
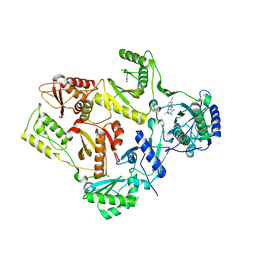 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-1 (PETT131A94) | | 分子名称: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-NITRILOMETHYL-PYRIDYL]-THIOUREA | | 著者 | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | 登録日 | 2000-01-13 | | 公開日 | 2000-03-20 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1E3G
 
 | | Human Androgen Receptor Ligand Binding in complex with the ligand metribolone (R1881) | | 分子名称: | (17BETA)-17-HYDROXY-17-METHYLESTRA-4,9,11-TRIEN-3-ONE, ANDROGEN RECEPTOR | | 著者 | Matias, P.M, Donner, P, Coelho, R, Thomaz, M, Peixoto, C, Macedo, S, Otto, N, Joschko, S, Scholz, P, Wegg, A, Basler, S, Schafer, M, Ruff, M, Egner, U, Carrondo, M.A. | | 登録日 | 2000-06-14 | | 公開日 | 2001-06-14 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural evidence for ligand specificity in the binding domain of the human androgen receptor. Implications for pathogenic gene mutations.
J. Biol. Chem., 275, 2000
|
|
1E64
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH LYS 75 REPLACED BY GLN (K75Q) | | 分子名称: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | 著者 | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | 登録日 | 2000-08-07 | | 公開日 | 2001-05-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Analysis of Interactions for Complex Formation between Ferredoxin-Nadp+ Reductase and its Protein Partners.
Proteins, 59, 2005
|
|
1ERZ
 
 | | CRYSTAL STRUCTURE OF N-CARBAMYL-D-AMINO ACID AMIDOHYDROLASE WITH A NOVEL CATALYTIC FRAMEWORK COMMON TO AMIDOHYDROLASES | | 分子名称: | N-CARBAMYL-D-AMINO ACID AMIDOHYDROLASE | | 著者 | Nakai, T, Hasegawa, T, Yamashita, E, Yamamoto, M, Kumasaka, T, Ueki, T, Nanba, H, Ikenaka, Y, Takahashi, S, Sato, M, Tsukihara, T. | | 登録日 | 2000-04-06 | | 公開日 | 2001-04-06 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of N-carbamyl-D-amino acid amidohydrolase with a novel catalytic framework common to amidohydrolases.
Structure Fold.Des., 8, 2000
|
|
1E62
 
 | | Ferredoxin:NADP+ reductase mutant with Lys 75 replaced by Arg (K75R) | | 分子名称: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | 著者 | Hermoso, J.A, Mayoral, T, Medina, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | 登録日 | 2000-08-07 | | 公開日 | 2001-05-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Analysis of Interactions for Complex Formation between Ferredoxin-Nadp+ Reductase and its Protein Partners.
Proteins, 59, 2005
|
|
1E63
 
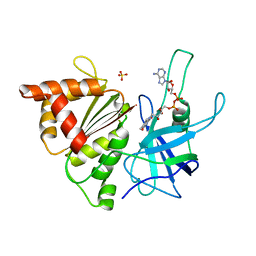 | | Ferredoxin:NADP+ Reductase Mutant with LYS 75 Replaced by SER (K75S) | | 分子名称: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | 著者 | Hermoso, J.A, Mayoral, T, Medina, M, Gomez-Moreno, C. | | 登録日 | 2000-08-07 | | 公開日 | 2001-05-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Analysis of Interactions for Complex Formation between Ferredoxin-Nadp+ Reductase and its Protein Partners.
Proteins, 59, 2005
|
|
1EQ8
 
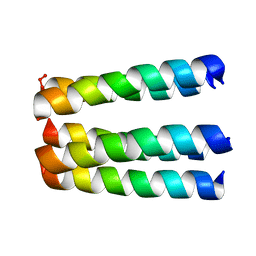 | | THREE-DIMENSIONAL STRUCTURE OF THE PENTAMERIC HELICAL BUNDLE OF THE ACETYLCHOLINE RECEPTOR M2 TRANSMEMBRANE SEGMENT | | 分子名称: | ACETYLCHOLINE RECEPTOR PROTEIN, HYDROXIDE ION | | 著者 | Marassi, F.M, Gesell, J.J, Kim, Y, Valente, A.P, Oblatt-Montal, M, Montal, M, Opella, S.J. | | 登録日 | 2000-04-03 | | 公開日 | 2000-04-26 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
4UU4
 
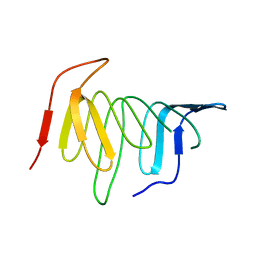 | | Crystal structure of LptH, the LptA homologous periplasmic component of the conserved lipopolysaccharide transport device from Pseudomonas aeruginosa | | 分子名称: | PERIPLASMIC LIPOPOLYSACCHARIDE TRANSPORT PROTEIN LPTH | | 著者 | Bollati, M, Villa, R, Gourlay, L.J, Barbiroli, A, Deho, G, Benedet, M, Polissi, A, Martorana, A, Sperandeo, P, Bolognesi, M, Nardini, M. | | 登録日 | 2014-07-24 | | 公開日 | 2015-03-25 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.751 Å) | | 主引用文献 | Crystal Structure of Lpth, the Periplasmic Component of the Lipopolysaccharide Transport Machinery from Pseudomonas Aeruginosa.
FEBS J., 282, 2015
|
|
