6O5J
 
 | | Crystal Structure of DAD2 bound to quinazolinone derivative | | Descriptor: | 1-(4-hydroxy-3-nitrophenyl)quinazoline-2,4(1H,3H)-dione, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hamiaux, C. | | Deposit date: | 2019-03-03 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Chemical synthesis and characterization of a new quinazolinedione competitive antagonist for strigolactone receptors with an unexpected binding mode.
Biochem.J., 476, 2019
|
|
6LI1
 
 | | Crystal structure of GPR52 ligand free form with flavodoxin fusion | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Flavodoxin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LI0
 
 | | Crystal structure of GPR52 in complex with agonist c17 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CITRATE ANION, Chimera of G-protein coupled receptor 52 and Flavodoxin, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
6LI2
 
 | | Crystal structure of GPR52 ligand free form with rubredoxin fusion | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera of G-protein coupled receptor 52 and Rubredoxin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Luo, Z.P, Lin, X, Xu, F, Han, G.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
5EW6
 
 | | Structure of ligand binding region of uPARAP at pH 7.4 without calcium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C-type mannose receptor 2, ... | | Authors: | Yuan, C, Huang, M. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of the ligand-binding region of uPARAP: effect of calcium ion binding
Biochem.J., 473, 2016
|
|
5ZM7
 
 | | Crystal structure of ORP1-ORD in complex with cholesterol at 3.4 A resolution | | Descriptor: | CHOLESTEROL, Oxysterol-binding protein-related protein 1 | | Authors: | Dong, J, Wang, J, Wu, J.W. | | Deposit date: | 2018-04-01 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P2/PI(3,4)P2.
Nat Commun, 10, 2019
|
|
5ZM5
 
 | | Crystal structure of human ORP1-ORD in complex with cholesterol at 2.6 A resolution | | Descriptor: | CHOLESTEROL, Oxysterol-binding protein-related protein 1 | | Authors: | Dong, J, Wang, J, Wu, J.W. | | Deposit date: | 2018-04-01 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric enhancement of ORP1-mediated cholesterol transport by PI(4,5)P2/PI(3,4)P2.
Nat Commun, 10, 2019
|
|
5HU4
 
 | | Cystal structure of listeria monocytogenes sortase A | | Descriptor: | Cysteine protease | | Authors: | Li, H. | | Deposit date: | 2016-01-27 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition of sortase A by chalcone prevents Listeria monocytogenes infection.
Biochem. Pharmacol., 106, 2016
|
|
7FIA
 
 | | Structure of AcrIF23 | | Descriptor: | AcrIF23 | | Authors: | Ren, J, Yue, F. | | Deposit date: | 2021-07-30 | | Release date: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural and mechanistic insights into the inhibition of type I-F CRISPR-Cas system by anti-CRISPR protein AcrIF23.
J.Biol.Chem., 298, 2022
|
|
7F5H
 
 | | The crystal structure of RBD-Nanobody complex, DL28 (SC4) | | Descriptor: | GLYCEROL, Nanobody DL28, PHOSPHATE ION, ... | | Authors: | Luo, Z.P, Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Characterization of a Neutralizing Nanobody With Broad Activity Against SARS-CoV-2 Variants.
Front Microbiol, 13, 2022
|
|
6LI3
 
 | | cryo-EM structure of GPR52-miniGs-NB35 | | Descriptor: | G-protein coupled receptor 52, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, M, Wang, N, Xu, F, Wu, J, Lei, M. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-18 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis of ligand recognition and self-activation of orphan GPR52.
Nature, 579, 2020
|
|
7WMF
 
 | | Threonyl-tRNA synthetase from Salmonella enterica in complex with an inhibitor | | Descriptor: | (2S,3R)-2-azanyl-N-[(1R)-2-[3-[6,7-bis(chloranyl)-4-oxidanylidene-quinazolin-3-yl]propanoylamino]-1-[3-[4-(trifluoromethyl)phenyl]phenyl]ethyl]-3-oxidanyl-butanamide, 1,2-ETHANEDIOL, Threonine--tRNA ligase, ... | | Authors: | Cai, Z, Chen, B, Yu, Y, Zhou, H. | | Deposit date: | 2022-01-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Proof-of-Concept of Triple-Site Inhibitors against Aminoacyl-tRNA Synthetases.
J.Med.Chem., 65, 2022
|
|
7WMI
 
 | | Threonyl-tRNA synthetase from Salmonella enterica in complex with an inhibitor | | Descriptor: | (2S,3R)-2-azanyl-N-[(1R)-2-[3-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)propanoylamino]-1-(3-phenylphenyl)ethyl]-3-oxidanyl-butanamide, 1,2-ETHANEDIOL, Threonine--tRNA ligase, ... | | Authors: | Cai, Z, Chen, B, Yu, Y, Zhou, H. | | Deposit date: | 2022-01-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Proof-of-Concept of Triple-Site Inhibitors against Aminoacyl-tRNA Synthetases.
J.Med.Chem., 65, 2022
|
|
7WM7
 
 | | Threonyl-tRNA synthetase from Salmonella enterica in complex with an inhibitor | | Descriptor: | (2S,3R)-2-azanyl-N-[5-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)pentyl]-3-oxidanyl-N-[[3-(1-oxidanylidene-2,3-dihydroisoindol-5-yl)phenyl]methyl]butanamide, Threonine--tRNA ligase, ZINC ION | | Authors: | Cai, Z, Chen, B, Yu, Y, Zhou, H. | | Deposit date: | 2022-01-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design, Synthesis, and Proof-of-Concept of Triple-Site Inhibitors against Aminoacyl-tRNA Synthetases.
J.Med.Chem., 65, 2022
|
|
6AP7
 
 | |
6AP6
 
 | | Crystal Structure of DAD2 in complex with tolfenamic acid | | Descriptor: | 2-[(3-chloro-2-methylphenyl)amino]benzoic acid, Probable strigolactone esterase DAD2 | | Authors: | Hamiaux, C. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibition of strigolactone receptors byN-phenylanthranilic acid derivatives: Structural and functional insights.
J. Biol. Chem., 293, 2018
|
|
6AP8
 
 | |
7EIV
 
 | | heterotetrameric glycyl-tRNA synthetase from Escherichia coli | | Descriptor: | GLYCINE, Glycine--tRNA ligase alpha subunit, Glycine--tRNA ligase beta subunit, ... | | Authors: | Ju, Y, Zhou, H. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | X-shaped structure of bacterial heterotetrameric tRNA synthetase suggests cryptic prokaryote functions and a rationale for synthetase classifications.
Nucleic Acids Res., 49, 2021
|
|
7F5G
 
 | | The crystal structure of RBD-Nanobody complex, DL4 (SA4) | | Descriptor: | ACETATE ION, GLYCEROL, Nanobody DL4, ... | | Authors: | Li, T, Lai, Y, Zhou, Y, Tan, J, Li, D. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Isolation, characterization, and structure-based engineering of a neutralizing nanobody against SARS-CoV-2.
Int.J.Biol.Macromol., 209, 2022
|
|
7XBR
 
 | | Crystal structure of phosphorylated AtMKK5 | | Descriptor: | Mitogen-activated protein kinase kinase 5 | | Authors: | Pei, C.J, Luo, Z.P, Wu, J.W, Wang, Z.X. | | Deposit date: | 2022-03-22 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the phosphorylated Arabidopsis MKK5 reveals activation mechanism of MAPK kinases.
Acta Biochim.Biophys.Sin., 54, 2022
|
|
7Y11
 
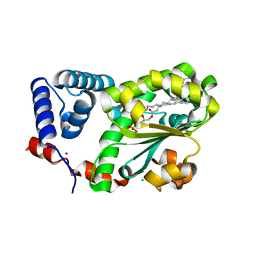 | | Crystal structure of AtSFH5-Sec14 in complex with egg PA | | Descriptor: | (2R)-1-(hexadecanoyloxy)-3-(phosphonooxy)propan-2-yl (9Z)-octadec-9-enoate, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Lu, Y.Q, Wang, X.Q, Luo, Z.P, Wu, J.W. | | Deposit date: | 2022-06-06 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Arabidopsis Sec14 proteins (SFH5 and SFH7) mediate interorganelle transport of phosphatidic acid and regulate chloroplast development.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Y10
 
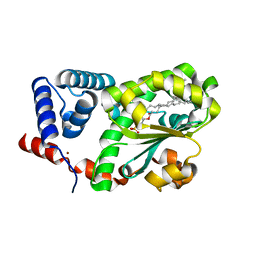 | | Crystal structure of AtSFH5-Sec14 in complex with DPPA | | Descriptor: | 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, NICKEL (II) ION, Phosphatidylinositol/phosphatidylcholine transfer protein SFH5 | | Authors: | Lu, Y.Q, Wang, X.Q, Luo, Z.P, Wu, J.W. | | Deposit date: | 2022-06-06 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Arabidopsis Sec14 proteins (SFH5 and SFH7) mediate interorganelle transport of phosphatidic acid and regulate chloroplast development.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8H2X
 
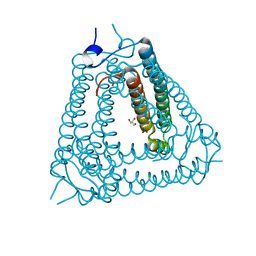 | | Structure of Acb2 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, p26 | | Authors: | Feng, Y, Cao, X.L. | | Deposit date: | 2022-10-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
8H2J
 
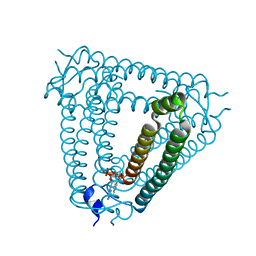 | | Structure of Acb2 complexed with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, p26 | | Authors: | Feng, Y, Cao, X.L. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
8H39
 
 | | Structure of Acb2 complexed with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, 1,2-ETHANEDIOL, p26 | | Authors: | Feng, Y, Cao, X.L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
