1Z29
 
 | | Crystal Structures of SULT1A2 and SULT1A1*3: Implications in the bioactivation of N-hydroxy-2-acetylamino fluorine (OH-AAF) | | Descriptor: | ACETIC ACID, ADENOSINE-3'-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Lu, J, Li, H, Liu, M.C, Zhang, J, Li, M, An, X, Chang, W. | | Deposit date: | 2005-03-07 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of SULT1A2 and SULT1A1 *3: insights into the substrate inhibition and the role of Tyr149 in SULT1A2.
Biochem.Biophys.Res.Commun., 396, 2010
|
|
1ZUB
 
 | | Solution Structure of the RIM1alpha PDZ Domain in Complex with an ELKS1b C-terminal Peptide | | Descriptor: | ELKS1b, Regulating synaptic membrane exocytosis protein 1 | | Authors: | Lu, J, Li, H, Wang, Y, Sudhof, T.C, Rizo, J. | | Deposit date: | 2005-05-30 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the RIM1alpha PDZ Domain in Complex with an ELKS1b C-terminal Peptide
J.Mol.Biol., 352, 2005
|
|
5GXO
 
 | | Discovery of a compound that activates SIRT3 to deacetylate Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Lu, J, Li, J, Wu, M, Wang, J, Xia, Q. | | Deposit date: | 2016-09-19 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A small molecule activator of SIRT3 promotes deacetylation and activation of manganese superoxide dismutase.
Free Radic. Biol. Med., 112, 2017
|
|
4MJ7
 
 | |
4URM
 
 | | Crystal Structure of Staph GyraseB 24kDa in complex with Kibdelomycin | | Descriptor: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA GYRASE SUBUNIT B | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4URL
 
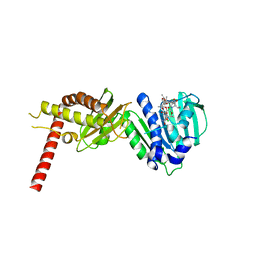 | | Crystal Structure of Staph ParE43kDa in complex with KBD | | Descriptor: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA TOPOISOMERASE IV, B SUBUNIT | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
4URN
 
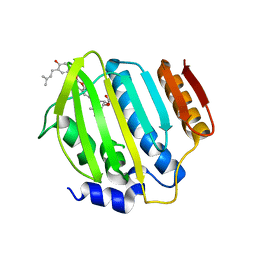 | | Crystal Structure of Staph ParE 24kDa in complex with Novobiocin | | Descriptor: | DNA TOPOISOMERASE IV, B SUBUNIT, NOVOBIOCIN | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-07-01 | | Release date: | 2014-07-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
2CJT
 
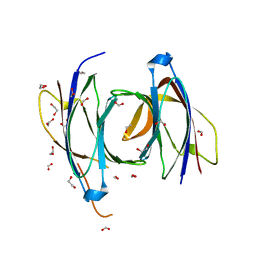 | | Structural Basis for a Munc13-1 Homodimer - Munc13-1 - RIM Heterodimer Switch: C2-domains as Versatile Protein-Protein Interaction Modules | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, UNC-13 HOMOLOG A | | Authors: | Lu, J, Machius, M, Dulubova, I, Dai, H, Sudhof, T.C, Tomchick, D.R, Rizo, J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Basis for a Munc13-1 Dimeric to Munc13-1/Rim Heterodimer Switch
Plos Biol., 4, 2006
|
|
2CJS
 
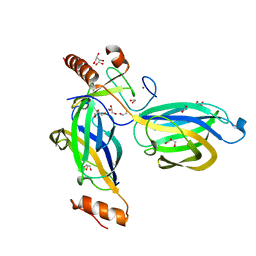 | | Structural Basis for a Munc13-1 Homodimer - Munc13-1 - RIM Heterodimer Switch: C2-domains as Versatile Protein-Protein Interaction Modules | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, REGULATING SYNAPTIC MEMBRANE EXOCYTOSIS PROTEIN 2, ... | | Authors: | Lu, J, Machius, M, Dulubova, I, Dai, H, Sudhof, T.C, Tomchick, D.R, Rizo, J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for a Munc13-1 Homodimer to Munc13-1/Rim Heterodimer Switch.
Plos Biol., 4, 2006
|
|
4X4M
 
 | | Structure of FcgammaRI in complex with Fc reveals the importance of glycan recognition for high affinity IgG binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lu, J, Sun, P.D. | | Deposit date: | 2014-12-03 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.485 Å) | | Cite: | Structure of Fc gamma RI in complex with Fc reveals the importance of glycan recognition for high-affinity IgG binding.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1KMD
 
 | | SOLUTION STRUCTURE OF THE VAM7P PX DOMAIN | | Descriptor: | Vacuolar morphogenesis protein VAM7 | | Authors: | Lu, J, Garcia, J, Dulubova, I, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-12-14 | | Release date: | 2002-06-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Vam7p PX domain.
Biochemistry, 41, 2002
|
|
5JQL
 
 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 Void of Bound Phospholipid from Saccharomyces Cerevisiae at 2.9 Angstroms Resolution | | Descriptor: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2016-05-05 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1
Commun Biol, 2020
|
|
5JQM
 
 | | Crystal Structure of Phosphatidic acid Transporter Ups1/Mdm35 Void of Bound Phospholipid from Saccharomyces Cerevisiae at 1.5 Angstroms Resolution | | Descriptor: | Protein UPS1, mitochondrial,Mitochondrial distribution and morphology protein 35 | | Authors: | Lu, J, Chan, K.C, Zhai, Y, Fan, J, Sun, F. | | Deposit date: | 2016-05-05 | | Release date: | 2017-07-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular mechanism of mitochondrial phosphatidate transfer by Ups1
Commun Biol, 2020
|
|
1Z28
 
 | | Crystal Structures of SULT1A2 and SULT1A1*3: Implications in the bioactivation of N-hydroxy-2-acetylamino fluorine (OH-AAF) | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Phenol-sulfating phenol sulfotransferase 1 | | Authors: | Lu, J, Li, H, Liu, M.C, Zhang, J, Li, M, An, X, Chang, W. | | Deposit date: | 2005-03-07 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of SULT1A2 and SULT1A1 *3: insights into the substrate inhibition and the role of Tyr149 in SULT1A2.
Biochem.Biophys.Res.Commun., 396, 2010
|
|
1MX8
 
 | |
1MX7
 
 | |
5TZY
 
 | | GPR40 in complex with AgoPAM AP8 and partial agonist MK-8666 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,3R)-3-cyclopropyl-3-[(2R)-2-(1-{(1S)-1-[5-fluoro-2-(trifluoromethoxy)phenyl]ethyl}piperidin-4-yl)-3,4-dihydro-2H-1-benzopyran-7-yl]-2-methylpropanoic acid, (5aR,6S,6aS)-3-({2',6'-dimethyl-4'-[3-(methylsulfonyl)propoxy][1,1'-biphenyl]-3-yl}methoxy)-5,5a,6,6a-tetrahydrocyclopropa[4,5]cyclopenta[1,2-c]pyridine-6-carboxylic acid, ... | | Authors: | Lu, J, Byrne, N, Patel, S, Sharma, S, Soisson, S.M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4URO
 
 | | Crystal Structure of Staph GyraseB 24kDa in complex with Novobiocin | | Descriptor: | DNA GYRASE SUBUNIT B, NOVOBIOCIN | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-07-01 | | Release date: | 2014-07-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
5K6E
 
 | |
5K38
 
 | |
5K74
 
 | |
1EII
 
 | | NMR STRUCTURE OF HOLO CELLULAR RETINOL-BINDING PROTEIN II | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN II, RETINOL | | Authors: | Lu, J, Lin, C.L, Tang, C, Ponder, J.W, Kao, J.L, Cistola, D.P, Li, E. | | Deposit date: | 2000-02-25 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Binding of retinol induces changes in rat cellular retinol-binding protein II conformation and backbone dynamics.
J.Mol.Biol., 300, 2000
|
|
8DIN
 
 | |
8DJ7
 
 | | The complex structure between human IgG1 Fc and its high affinity receptor FcgRI H174R variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin gamma Fc receptor I, Ig gamma-1 Fc chain, ... | | Authors: | Lu, J, Sun, P.D. | | Deposit date: | 2022-06-30 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Fc gamma RI FG-loop functions as a pH sensitive switch for IgG binding and release.
Front Immunol, 14, 2023
|
|
6WPQ
 
 | | GNYNVF from hnRNPA2-low complexity domain segment, residues 286-291, D290V variant | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A2 | | Authors: | Lu, J, Cao, Q, Hughes, M.P, Sawaya, M.R, Boyer, D.R, Cascio, D, Eisenberg, D.S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid.
Nat Commun, 11, 2020
|
|
