1IM9
 
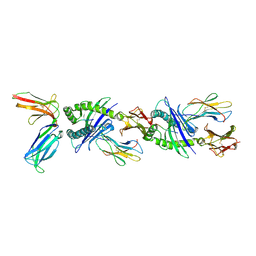 | | Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1 bound to its MHC ligand HLA-Cw4 | | Descriptor: | Beta-2 microglobulin, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, CW-4 CW*0401 ALPHA CHAIN, ... | | Authors: | Fan, Q.R, Long, E.O, Wiley, D.C. | | Deposit date: | 2001-05-10 | | Release date: | 2001-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex.
Nat.Immunol., 2, 2001
|
|
1NKR
 
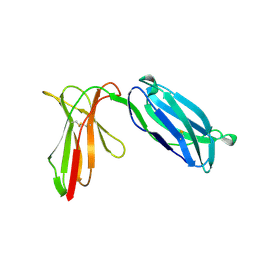 | | INHIBITORY RECEPTOR (P58-CL42) FOR HUMAN NATURAL KILLER CELLS | | Descriptor: | P58-CL42 KIR | | Authors: | Fan, Q.R, Mosyak, L, Winter, C.C, Wagtmann, N, Long, E.O, Wiley, D.C. | | Deposit date: | 1998-06-24 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the inhibitory receptor for human natural killer cells resembles haematopoietic receptors.
Nature, 389, 1997
|
|
6ULR
 
 | |
6ULI
 
 | |
7SU9
 
 | |
6ULK
 
 | |
6ULN
 
 | |
6UON
 
 | |
1NLR
 
 | | ENDO-1,4-BETA-GLUCANASE CELB2, CELLULASE, NATIVE STRUCTURE | | Descriptor: | ENDO-1,4-BETA-GLUCANASE | | Authors: | Sulzenbacher, G, Dupont, C, Davies, G.J. | | Deposit date: | 1997-10-27 | | Release date: | 1998-11-25 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Streptomyces lividans family 12 endoglucanase: construction of the catalytic cre, expression, and X-ray structure at 1.75 A resolution.
Biochemistry, 36, 1997
|
|
