6V2S
 
 | | Crystal Structure of chromodomain of MPP8 in complex with inhibitor UNC3866 | | Descriptor: | M-phase phosphoprotein 8, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V2R
 
 | | Crystal Structure of chromodomain of CBX7 mutant V13A in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 7, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V2D
 
 | | Crystal Structure of chromodomain of CDYL2 in complex with inhibitor UNC3866 | | Descriptor: | Chromodomain Y-like protein 2, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
1LXT
 
 | | STRUCTURE OF PHOSPHOTRANSFERASE PHOSPHOGLUCOMUTASE FROM RABBIT | | Descriptor: | CADMIUM ION, PHOSPHOGLUCOMUTASE (DEPHOSPHO FORM), SULFATE ION | | Authors: | Ray Junior, W.J, Baranidharan, S, Liu, Y. | | Deposit date: | 1996-07-28 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of rabbit muscle phosphoglucomutase refined at 2.4 A resolution.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
6UE6
 
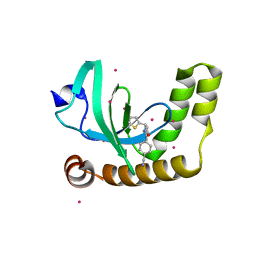 | | PWWP1 domain of NSD2 in complex with MR837 | | Descriptor: | 4-cyano-N-cyclopropyl-N-[(thiophen-2-yl)methyl]benzamide, Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, De Freitas, R.F, Schapira, M, Brown, P.J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-09-20 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Small-Molecule Antagonists of the PWWP Domain of NSD2.
J.Med.Chem., 64, 2021
|
|
6UPL
 
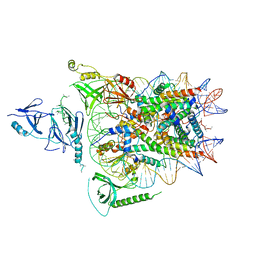 | | Structure of FACT_subnucleosome complex 2 | | Descriptor: | DNA (79-mer), FACT complex subunit SPT16, FACT complex subunit SSRP1, ... | | Authors: | Zhou, K, Tan, Y.Z, Wei, H, Liu, Y, Carragher, B, Potter, C, Luger, K. | | Deposit date: | 2019-10-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | FACT caught in the act of manipulating the nucleosome.
Nature, 577, 2020
|
|
6UPK
 
 | | Structure of FACT_subnucleosome complex 1 | | Descriptor: | DNA (79-mer), FACT complex subunit SPT16, FACT complex subunit SSRP1, ... | | Authors: | Zhou, K, Tan, Y.Z, Wei, H, Liu, Y, Carragher, B, Potter, C, Luger, K. | | Deposit date: | 2019-10-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | FACT caught in the act of manipulating the nucleosome.
Nature, 577, 2020
|
|
4QQG
 
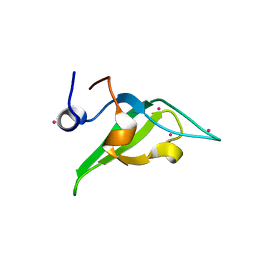 | | Crystal structure of an N-terminal HTATIP fragment | | Descriptor: | Histone acetyltransferase KAT5, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Wernimont, A.K, Dobrovetsky, E, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and histone binding studies of the chromo barrel domain of TIP60.
FEBS Lett., 592, 2018
|
|
6C9I
 
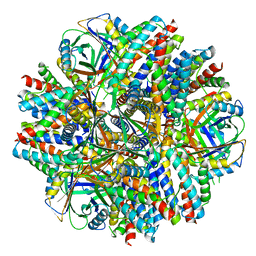 | | Single-Particle reconstruction of DARP14 - A designed protein scaffold displaying ~17kDa DARPin proteins - Scaffold | | Descriptor: | DARP14 - Subunit A with DARPin, DARP14 - Subunit B | | Authors: | Gonen, S, Liu, Y, Yeates, T.O, Gonen, T. | | Deposit date: | 2018-01-26 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Near-atomic cryo-EM imaging of a small protein displayed on a designed scaffolding system.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
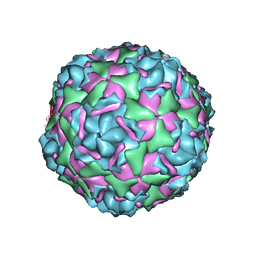
6CV5
 
 | |
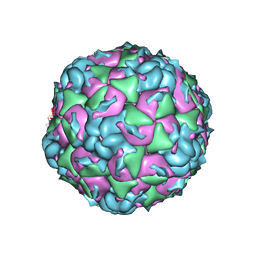
6CV4
 
 | |
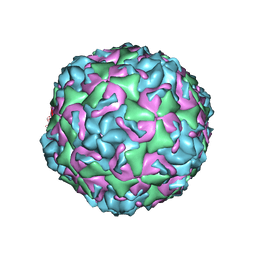
6CV2
 
 | |
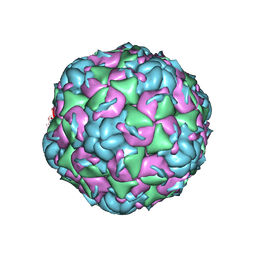
6CV3
 
 | |
6CV1
 
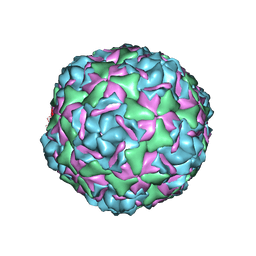 | |
6CVB
 
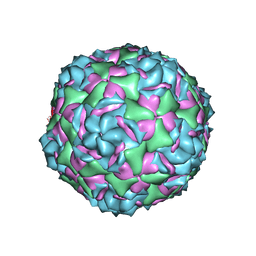 | |
6D5F
 
 | | Cryo-EM reconstruction of membrane-enveloped filamentous virus SFV1 (Sulfolobus filamentous virus 1) | | Descriptor: | DNA (336-MER), Fimbrial protein | | Authors: | Wang, F, Osinski, T, Liu, Y, Krupovic, M, Prangishvili, D, Egelman, E.H. | | Deposit date: | 2018-04-19 | | Release date: | 2018-08-29 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural conservation in a membrane-enveloped filamentous virus infecting a hyperthermophilic acidophile.
Nat Commun, 9, 2018
|
|
5W3M
 
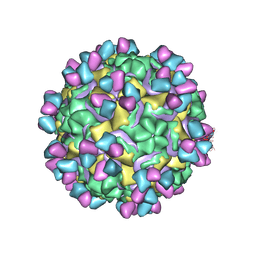 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:1, full particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W3O
 
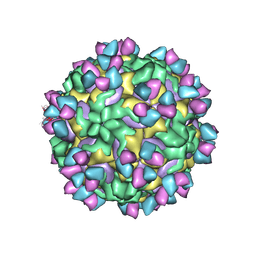 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:3, empty particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W3L
 
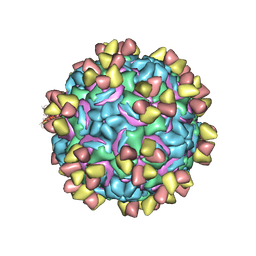 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (4 degrees Celsius, molar ratio 1:3, full particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W3E
 
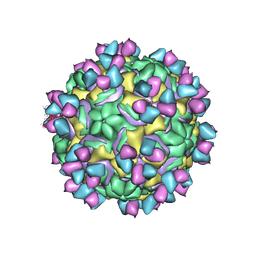 | | CryoEM structure of rhinovirus B14 in complex with C5 Fab (33 degrees Celsius, molar ratio 1:3, full particle) | | Descriptor: | C5 antibody variable heavy domain, C5 antibody variable light domain, viral protein 1, ... | | Authors: | Liu, Y, Dong, Y, Rossmann, M.G. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Antibody-induced uncoating of human rhinovirus B14.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5IN1
 
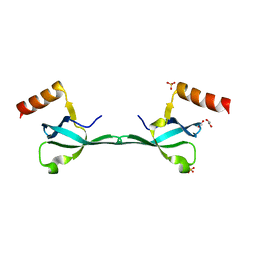 | | Crystal Structure of the MRG701 chromodomain | | Descriptor: | 1,2-ETHANEDIOL, MRG701, SULFATE ION | | Authors: | Huang, Y, Liu, Y. | | Deposit date: | 2016-03-07 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on MRG701 chromodomain reveal a novel dimerization interface of MRG proteins in green plants
Protein Cell, 7, 2016
|
|
2QAF
 
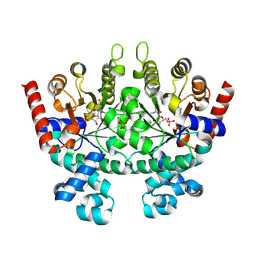 | | Crystal structure of Plasmodium falciparum orotidine 5'-phosphate decarboxylase covalently modified by 6-iodo-UMP | | Descriptor: | Orotidine 5' monophosphate decarboxylase, SULFATE ION, URIDINE-5'-MONOPHOSPHATE | | Authors: | Liu, Y, Lau, W, Bello, A.M, Kotra, L.P, Hui, R, Pai, E.F. | | Deposit date: | 2007-06-15 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-activity relationships of C6-uridine derivatives targeting plasmodia orotidine monophosphate decarboxylase
J.Med.Chem., 51, 2008
|
|
3PMG
 
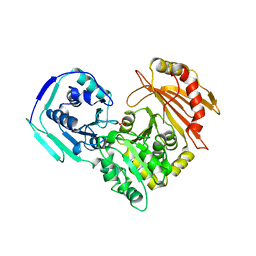 | |
6IH5
 
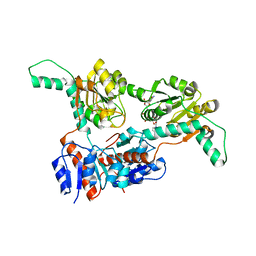 | | Crystal structure of Phosphite Dehydrogenase mutant I151R/P176E from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine dinucleotide | | Descriptor: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Song, X, Feng, Y, Liu, Y, Zhao, Z. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH4
 
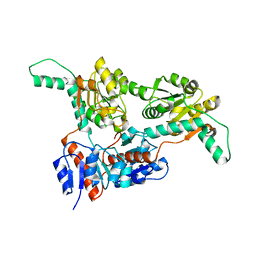 | |
