5T1I
 
 | | CBX3 chromo shadow domain in complex with histone H3 peptide | | Descriptor: | Chromobox protein homolog 3, Histone H3.1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-08-19 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Peptide recognition by heterochromatin protein 1 (HP1) chromoshadow domains revisited: Plasticity in the pseudosymmetric histone binding site of human HP1.
J. Biol. Chem., 292, 2017
|
|
3LKX
 
 | | Human nac dimerization domain | | Descriptor: | Nascent polypeptide-associated complex subunit alpha, Transcription factor BTF3 | | Authors: | Liu, Y, Hu, Y, Li, X, Niu, L, Teng, M. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the human nascent polypeptide-associated complex domain reveals a nucleic acid-binding region on the NACA subunit
Biochemistry, 49, 2010
|
|
1DQO
 
 | | Crystal structure of the cysteine rich domain of mannose receptor complexed with Acetylgalactosamine-4-sulfate | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, MANNOSE RECEPTOR | | Authors: | Liu, Y, Chirino, A.J, Misulovin, Z, Leteux, C, Feizi, T, Nussenzweig, M.C, Bjorkman, P.J. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the cysteine-rich domain of mannose receptor complexed with a sulfated carbohydrate ligand.
J.Exp.Med., 191, 2000
|
|
1JH5
 
 | | Crystal Structure of sTALL-1 of TNF family ligand | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 13B | | Authors: | Liu, Y, Xu, L, Opalka, N, Shu, H.-B, Zhang, G. | | Deposit date: | 2001-06-27 | | Release date: | 2002-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of sTALL-1 reveals a virus-like assembly of TNF family ligands.
Cell(Cambridge,Mass.), 108, 2002
|
|
1FWV
 
 | |
1FWU
 
 | |
1DQG
 
 | | CRYSTAL STRUCTURE OF THE CYSTEINE RICH DOMAIN OF MANNOSE RECEPTOR | | Descriptor: | MANNOSE RECEPTOR, SULFATE ION | | Authors: | Liu, Y, Chirino, A.J, Misulovin, Z, Leteux, C, Feizi, T, Nussenzweig, M.C, Bjorkman, P.J. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the cysteine-rich domain of mannose receptor complexed with a sulfated carbohydrate ligand.
J.Exp.Med., 191, 2000
|
|
7N8N
 
 | | Melbournevirus nucleosome like particle | | Descriptor: | DNA (147-MER), Histone H2B-H2A doublet, Histone H4-H3 doublet | | Authors: | Liu, Y, Toner, C.M, Zhou, K, Bowerman, S, Luger, K. | | Deposit date: | 2021-06-15 | | Release date: | 2021-08-04 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Virus-encoded histone doublets are essential and form nucleosome-like structures.
Cell, 184, 2021
|
|
3A9B
 
 | | CcCel6C, a glycoside hydrolase family 6 enzyme, complexed with cellobiose | | Descriptor: | Cellobiohydrolase, MAGNESIUM ION, beta-D-glucopyranose, ... | | Authors: | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
3ABX
 
 | | CcCel6C, a glycoside hydrolase family 6 enzyme, complexed with p-nitrophenyl beta-D-cellotrioside | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranosyl-(1->4)-beta-D-glucopyranoside, Cellobiohydrolase, MAGNESIUM ION | | Authors: | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-12-24 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
6NHT
 
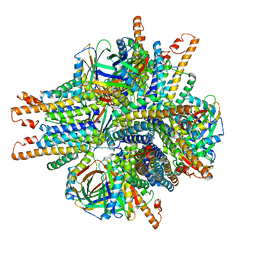 | |
6NHV
 
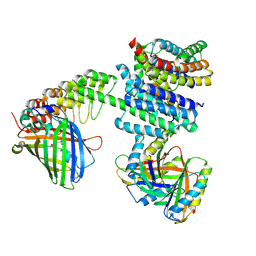 | |
4WM7
 
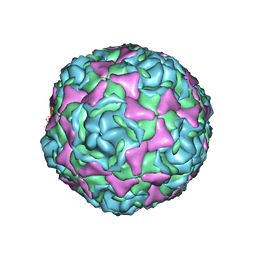 | | Crystal Structure of Human Enterovirus D68 in Complex with Pleconaril | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, VP1, VP2, ... | | Authors: | Liu, Y, Sheng, J, Fokine, A, Meng, G, Long, F, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2014-10-08 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Virus structure. Structure and inhibition of EV-D68, a virus that causes respiratory illness in children.
Science, 347, 2015
|
|
4WM8
 
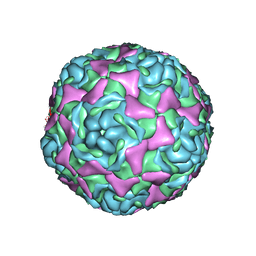 | | Crystal Structure of Human Enterovirus D68 | | Descriptor: | DECANOIC ACID, VP1, VP2, ... | | Authors: | Liu, Y, Sheng, J, Fokine, A, Meng, G, Long, F, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2014-10-08 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Virus structure. Structure and inhibition of EV-D68, a virus that causes respiratory illness in children.
Science, 347, 2015
|
|
7TNC
 
 | | M13F/G116F Pseudomonas aeruginosa azurin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Azurin, CHLORIDE ION, ... | | Authors: | Liu, Y, Lu, Y. | | Deposit date: | 2022-01-20 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Basis for the Effects of Phenylalanine on Tuning the Reduction Potential of Type 1 Copper in Azurin.
Inorg.Chem., 62, 2023
|
|
4BL6
 
 | | Bicaudal-D uses a parallel, homodimeric coiled coil with heterotypic registry to co-ordinate recruitment of cargos to dynein | | Descriptor: | ARGININE, PROTEIN BICAUDAL D | | Authors: | Liu, Y, Salter, H.K, Holding, A.N, Johnson, C.M, Stephens, E, Lukavsky, P.J, Walshaw, J, Bullock, S.L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-06-12 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Bicaudal-D Uses a Parallel, Homodimeric Coiled Coil with Heterotypic Registry to Coordinate Recruitment of Cargos to Dynein
Genes Dev., 27, 2013
|
|
7KDP
 
 | | HCMV prefusion gB in complex with fusion inhibitor WAY-174865 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein B, ... | | Authors: | Liu, Y, Heim, P.K, Che, Y, Chi, X, Qiu, X, Han, S, Dormitzer, P.R, Yang, X. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Prefusion structure of human cytomegalovirus glycoprotein B and structural basis for membrane fusion.
Sci Adv, 7, 2021
|
|
7KDD
 
 | | HCMV postfusion gB in complex with SM5-1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein B, ... | | Authors: | Liu, Y, Heim, P.K, Che, Y, Chi, X, Qiu, X, Han, S, Dormitzer, P.R, Yang, X. | | Deposit date: | 2020-10-08 | | Release date: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Prefusion structure of human cytomegalovirus glycoprotein B and structural basis for membrane fusion.
Sci Adv, 7, 2021
|
|
1WQ6
 
 | | The tetramer structure of the nervy homolgy two (NHR2) domain of AML1-ETO is critical for AML1-ETO'S activity | | Descriptor: | AML1-ETO | | Authors: | Liu, Y, Cheney, M.D, Chruszcz, M, Lukasik, S.M, Hartman, K.L, Laue, T.M, Dauter, Z, Minor, W, Speck, N.A, Bushweller, J.H. | | Deposit date: | 2004-09-23 | | Release date: | 2005-10-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity
Cancer Cell, 9, 2006
|
|
5F3W
 
 | | Structure of the ATPrS-Mre11/Rad50-DNA complex | | Descriptor: | 27-MER DNA, DNA double-strand break repair Rad50 ATPase,DNA double-strand break repair Rad50 ATPase, DNA double-strand break repair protein Mre11, ... | | Authors: | Liu, Y. | | Deposit date: | 2015-12-03 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex
Embo J., 35, 2016
|
|
1YXO
 
 | | Crystal Structure of pyridoxal phosphate biosynthetic protein PdxA PA0593 | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase 1, ETHANOL, MAGNESIUM ION | | Authors: | Liu, Y, Xu, X, Dong, A, Kudritskam, M, Savchenko, A, Pai, E.F, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-05 | | Last modified: | 2011-10-05 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of pyridoxal phosphate biosynthetic protein PdxA PA0593
To be Published
|
|
5DNY
 
 | | Structure of the ATPrS-Mre11/Rad50-DNA complex | | Descriptor: | DNA (27-MER), DNA double-strand break repair Rad50 ATPase,DNA double-strand break repair Rad50 ATPase, DNA double-strand break repair protein Mre11, ... | | Authors: | Liu, Y. | | Deposit date: | 2015-09-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex.
Embo J., 35, 2016
|
|
1YQE
 
 | | Crystal Structure of Conserved Protein of Unknown Function AF0625 | | Descriptor: | Hypothetical UPF0204 protein AF0625, PYROPHOSPHATE 2- | | Authors: | Liu, Y, Skarina, T, Dong, A, Kudritskam, M, Savchenko, A, Pai, E.F, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-01 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein AF0625
To be Published
|
|
3MW7
 
 | |
3N2M
 
 | | Crystal structure of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase complexed with 5-fluoro-6-amino-UMP | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-amino-5-fluorouridine 5'-(dihydrogen phosphate), ... | | Authors: | Liu, Y, Kotra, L.P, Pai, E.F. | | Deposit date: | 2010-05-18 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase complexed with 5-fluoro-6-amino-UMP
To be Published
|
|
