1XZ3
 
 | | Complex of apoferritin with isoflurane | | 分子名称: | 1-CHLORO-2,2,2-TRIFLUOROETHYL DIFLUOROMETHYL ETHER, CADMIUM ION, Ferritin light chain | | 著者 | Liu, R, Loll, P.J, Eckenhoff, R.G. | | 登録日 | 2004-11-11 | | 公開日 | 2005-05-10 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for high-affinity volatile anesthetic binding in a natural 4-helix bundle protein.
Faseb J., 19, 2005
|
|
1XZ1
 
 | | Complex of halothane with apoferritin | | 分子名称: | 2-BROMO-2-CHLORO-1,1,1-TRIFLUOROETHANE, CADMIUM ION, Ferritin light chain | | 著者 | Liu, R, Loll, P.J, Eckenhoff, R.G. | | 登録日 | 2004-11-11 | | 公開日 | 2005-05-10 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural basis for high-affinity volatile anesthetic binding in a natural 4-helix bundle protein.
Faseb J., 19, 2005
|
|
1QS9
 
 | |
1QTD
 
 | |
1QS5
 
 | |
1QSB
 
 | |
1QTH
 
 | |
1QTC
 
 | |
1QTB
 
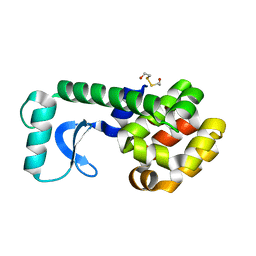 | |
5YXI
 
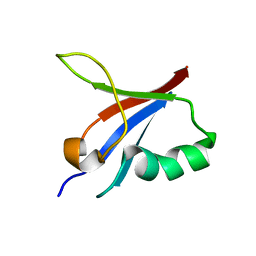 | | Designed protein dRafX6 | | 分子名称: | Designed protein dRafX6 | | 著者 | Liu, R. | | 登録日 | 2017-12-05 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo sequence redesign of a functional Ras-binding domain globally inverted the surface charge distribution and led to extreme thermostability.
Biotechnol.Bioeng., 118, 2021
|
|
3NDP
 
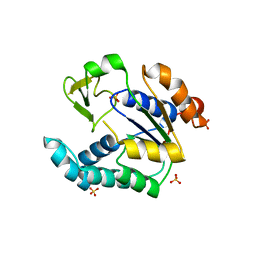 | | Crystal structure of human AK4(L171P) | | 分子名称: | Adenylate kinase isoenzyme 4, SULFATE ION | | 著者 | Liu, R, Wang, Y, Wei, Z, Gong, W. | | 登録日 | 2010-06-07 | | 公開日 | 2010-06-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of human adenylate kinase 4 (L171P) suggests the role of hinge region in protein domain motion
Biochem.Biophys.Res.Commun., 379, 2009
|
|
3U90
 
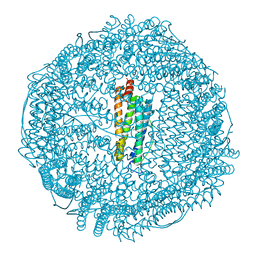 | | apoferritin: complex with SDS | | 分子名称: | CADMIUM ION, DODECYL SULFATE, Ferritin light chain | | 著者 | Liu, R, Bu, W, Xi, J, Mortazavi, S.R, Cheung-Lau, J.C, Dmochowski, I.J, Loll, P.J. | | 登録日 | 2011-10-17 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Beyond the detergent effect: a binding site for sodium dodecyl sulfate (SDS) in mammalian apoferritin.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8GZ3
 
 | |
8HBE
 
 | |
8HBF
 
 | |
8HBH
 
 | |
3GEX
 
 | |
7VSQ
 
 | |
4H48
 
 | | 1.45 angstrom CyPet Structure at pH7.0 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Green fluorescent protein | | 著者 | Hu, X.-J, Liu, R. | | 登録日 | 2012-09-17 | | 公開日 | 2013-09-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure insight of the fluorescent state of CyPet
To be Published
|
|
4H47
 
 | |
7D9R
 
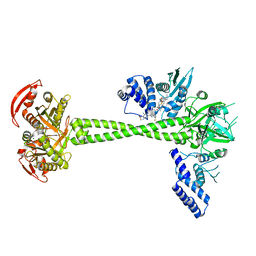 | | Structure of huamn soluble guanylate cyclase in the riociguat and NO-bound state | | 分子名称: | Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, MAGNESIUM ION, ... | | 著者 | Chen, L, Liu, R, Kang, Y. | | 登録日 | 2020-10-14 | | 公開日 | 2021-08-11 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Activation mechanism of human soluble guanylate cyclase by stimulators and activators.
Nat Commun, 12, 2021
|
|
7D9T
 
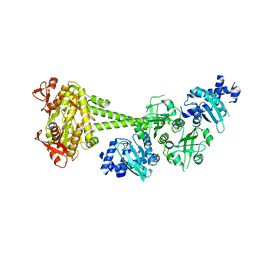 | |
7D9U
 
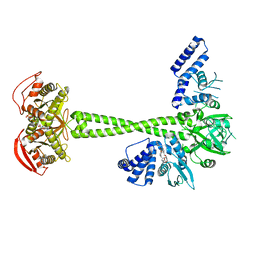 | | Structure of human soluble guanylate cyclase in the cinciguat-bound activated state | | 分子名称: | 4-({(4-carboxybutyl)[2-(2-{[4-(2-phenylethyl)benzyl]oxy}phenyl)ethyl]amino}methyl)benzoic acid, Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, ... | | 著者 | Chen, L, Liu, R, Kang, Y. | | 登録日 | 2020-10-14 | | 公開日 | 2021-08-11 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Activation mechanism of human soluble guanylate cyclase by stimulators and activators.
Nat Commun, 12, 2021
|
|
7D9S
 
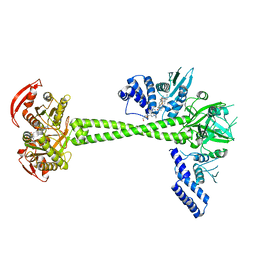 | | Structure of huamn soluble guanylate cyclase in the YC1 and NO-bound state | | 分子名称: | Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, MAGNESIUM ION, ... | | 著者 | Chen, L, Liu, R, Kang, Y. | | 登録日 | 2020-10-14 | | 公開日 | 2021-08-11 | | 最終更新日 | 2021-10-13 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Activation mechanism of human soluble guanylate cyclase by stimulators and activators.
Nat Commun, 12, 2021
|
|
6LQE
 
 | |
