2NOC
 
 | | Solution Structure of Putative periplasmic protein: Northest Structural Genomics Target StR106 | | Descriptor: | Putative periplasmic protein | | Authors: | Zhang, Q, Liu, G, Wang, H, Nwosu, C, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Putative periplasmic protein: Northest Structural Genomics Target StR106
To be Published
|
|
2NWT
 
 | | NMR Structure of Protein UPF0165 protein AF_2212 from Archaeoglobus Fulgidus; Northeast Structural Genomics Consortium Target GR83 | | Descriptor: | UPF0165 protein AF_2212 | | Authors: | Singarapu, K.K, Sukumaran, D.K, Parish, D, Atreya, H.S, Liu, G, Eletsky, A, Chen, C.X, Jiang, M, Cunningham, K, Xiao, R, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-16 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Protein Y2212_ARCFU from Archaeoglobus Fulgidus; Northeast Structural Genomics Consortium Target GR83
To be Published
|
|
4AL0
 
 | | Crystal structure of Human PS-1 | | Descriptor: | GLUTATHIONE, PALMITIC ACID, PROSTAGLANDIN E SYNTHASE, ... | | Authors: | Sjogren, T, Nord, J, Ek, M, Johansson, P, Liu, G, Geschwindner, S. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal Structure of Microsomal Prostaglandin E2 Synthase Provides Insight Into Diversity in the Mapeg Superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AL1
 
 | | Crystal structure of Human PS-1 GSH-analog complex | | Descriptor: | L-gamma-glutamyl-S-(2-biphenyl-4-yl-2-oxoethyl)-L-cysteinylglycine, PALMITIC ACID, PROSTAGLANDIN E SYNTHASE, ... | | Authors: | Sjogren, T, Nord, J, Ek, M, Johansson, P, Liu, G, Geschwindner, S. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Microsomal Prostaglandin E2 Synthase Provides Insight Into Diversity in the Mapeg Superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6JXM
 
 | | Crystal Structure of phi29 pRNA domain II | | Descriptor: | BARIUM ION, MAGNESIUM ION, RNA (97-mer) | | Authors: | Cai, R, Price, I.R, Ding, F, Wu, F, Chen, T, Zhang, Y, Liu, G, Jardine, P.J, Lu, C, Ke, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | ATP/ADP modulates gp16-pRNA conformational change in the Phi29 DNA packaging motor.
Nucleic Acids Res., 47, 2019
|
|
5TTT
 
 | | Sparse-restraint solution NMR structure of micelle-solubilized cytosolic amino terminal domain of C. elegans mechanosensory ion channel MEC-4 refined by restrained Rosetta | | Descriptor: | Degenerin mec-4 | | Authors: | Everett, J.K, Liu, G, Mao, B, Driscoll, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sparse-restraint solution NMR structure of micelle-solubilized
cytosolic amino terminal domain of C. elegans mechanosensory ion channel
MEC-4 refined by restrained Rosetta
To Be Published
|
|
6NS8
 
 | | RDC-refined SOLUTION NMR STRUCTURE OF PROTEIN PF2048.1 | | Descriptor: | Uncharacterized protein | | Authors: | Daigham, N.S, Liu, G, Swapna, G.V.T, Cole, C, Valafar, H, Montelione, G.T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | REDCRAFT: A Computational Platform Using Residual Dipolar Coupling NMR Data for Determining Structures of Perdeuterated Proteins Without NOEs
To Be Published
|
|
8IZ4
 
 | | Lysophosphatidylserine receptor GPR34-Gi complex | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Gong, W, Liu, G, Li, X, Zhang, X. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural mechanisms of ligand binding and signaling in lysophosphatidylserine receptors
To Be Published
|
|
2K2B
 
 | |
2JZT
 
 | | Solution NMR structure of Q8ZP25_SALTY from Salmonella typhimurium. Northeast Structural Genomics Consortium target StR70 | | Descriptor: | Putative thiol-disulfide isomerase and thioredoxin | | Authors: | Parish, D, Liu, G, Shen, Y, Ho, C, Cunningham, K, Xiao, R, Swapna, G.V.T, Acton, T, Bansal, S, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Protein chaperones Q8ZP25_SALTY from Salmonella typhimurium and HYAE_ECOLI from Escherichia coli exhibit thioredoxin-like structures despite lack of canonical thioredoxin active site sequence motif.
J.STRUCT.FUNCT.GENOM., 9, 2008
|
|
2JNA
 
 | | Solution NMR Structure of Salmonella typhimurium LT2 Secreted Protein STM0082: Northeast Structural Genomics Consortium Target StR109 | | Descriptor: | Putative secreted protein | | Authors: | Eletsky, A, Parish, D, Liu, G, Sukumaran, D, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-31 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Salmonella typhimurium LT2 Secreted Protein STM0082: Northeast Structural Genomics Consortium Target StR109
To be Published
|
|
2LNI
 
 | | Solution NMR Structure of Stress-induced-phosphoprotein 1 STI1 from Homo sapiens, Northeast Structural Genomics Consortium Target HR4403E | | Descriptor: | Stress-induced-phosphoprotein 1 | | Authors: | Tang, Y, Liu, G, Hamilton, K, Ciccosanti, C, Shastry, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-12-28 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target HR4403E
To be Published
|
|
2M89
 
 | | Solution structure of the Aha1 dimer from Colwellia psychrerythraea | | Descriptor: | Aha1 domain protein | | Authors: | Rossi, P, Sgourakis, N.G, Shi, L, Liu, G, Barbieri, C.M, Lee, H, Grant, T.D, Luft, J.R, Xiao, R, Acton, T.B, Montelione, G.T, Snell, E.H, Baker, D, Lange, O.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-09 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | A hybrid NMR/SAXS-based approach for discriminating oligomeric protein interfaces using Rosetta.
Proteins, 83, 2015
|
|
7JYZ
 
 | | Solution NMR structure and dynamics of human Brd3 ET in complex with MLV IN CTD | | Descriptor: | Bromodomain-containing protein 3, Integrase | | Authors: | Aiyer, S, Liu, G, Swapna, G.V.T, Hao, J, Ma, L.C, Roth, M.J, Montelione, G.T. | | Deposit date: | 2020-09-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
7K3S
 
 | |
7WBO
 
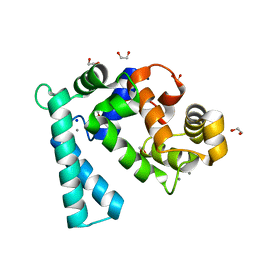 | | Crystal structure of Sarcoplasmic Calcium-Binding Protein from Scylla paramamosain | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | Authors: | Chen, Y, Jin, T, Liu, G. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure Analysis of Sarcoplasmic-Calcium-Binding Protein: An Allergen in Scylla paramamosain.
J.Agric.Food Chem., 71, 2023
|
|
3EVX
 
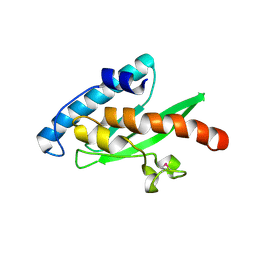 | | Crystal structure of the human E2-like ubiquitin-fold modifier conjugating enzyme 1 (Ufc1). Northeast Structural Genomics Consortium target HR41 | | Descriptor: | THIOCYANATE ION, Ufm1-conjugating enzyme 1 | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-13 | | Release date: | 2008-10-21 | | Last modified: | 2023-01-04 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | NMR and X-RAY structures of human E2-like ubiquitin-fold modifier conjugating enzyme 1 (UFC1) reveal structural and functional conservation in the metazoan UFM1-UBA5-UFC1 ubiquination pathway.
J.STRUCT.FUNCT.GENOM., 10, 2009
|
|
8JIY
 
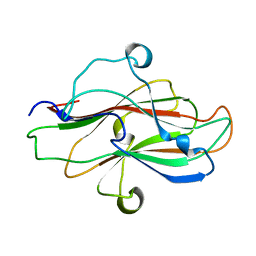 | | A carbohydrate binding domain of a putative chondroitinase | | Descriptor: | DUF4955 domain-containing protein | | Authors: | Liu, G.C, Chang, Y.G. | | Deposit date: | 2023-05-29 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and structural characterization of a novel chondroitin sulfate-specific carbohydrate-binding module: The first member of a new family, CBM100.
Int.J.Biol.Macromol., 255, 2024
|
|
6IG9
 
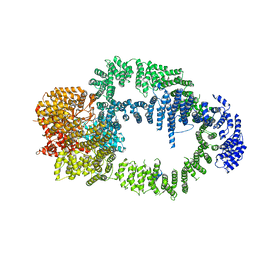 | |
7XM2
 
 | | Crystal structure of Keap1 Kelch domain (residues 322-609) in complex with NXPZ-2 | | Descriptor: | 2-[(4-aminophenyl)sulfonyl-[4-[(2-azanyl-2-oxidanylidene-ethyl)-(4-methoxyphenyl)sulfonyl-amino]naphthalen-1-yl]amino]ethanamide, Kelch-like ECH-associated protein 1 | | Authors: | Xu, K. | | Deposit date: | 2022-04-24 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallography-Guided Optimizations of the Keap1-Nrf2 Inhibitors on the Solvent Exposed Region: From Symmetric to Asymmetric Naphthalenesulfonamides.
J.Med.Chem., 65, 2022
|
|
7XM4
 
 | | Crystal structure of Keap1 Kelch domain (residues 322-609) in complex with 6e | | Descriptor: | Kelch-like ECH-associated protein 1, N-[4-[(2-azanyl-2-oxidanylidene-ethyl)-[4-[(2-azanyl-2-oxidanylidene-ethyl)-(4-methoxyphenyl)sulfonyl-amino]naphthalen-1-yl]sulfamoyl]phenyl]-2-(4-ethylpiperazin-1-yl)ethanamide | | Authors: | Xu, K. | | Deposit date: | 2022-04-24 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallography-Guided Optimizations of the Keap1-Nrf2 Inhibitors on the Solvent Exposed Region: From Symmetric to Asymmetric Naphthalenesulfonamides.
J.Med.Chem., 65, 2022
|
|
7XM3
 
 | | Crystal structure of Keap1 Kelch domain (residues 322-609) in complex with 6k | | Descriptor: | Kelch-like ECH-associated protein 1, N-[4-[(2-azanyl-2-oxidanylidene-ethyl)-[4-[(2-azanyl-2-oxidanylidene-ethyl)-(4-methoxyphenyl)sulfonyl-amino]naphthalen-1-yl]sulfamoyl]phenyl]-3-(4-ethylpiperazin-1-yl)propanamide | | Authors: | Xu, K. | | Deposit date: | 2022-04-24 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallography-Guided Optimizations of the Keap1-Nrf2 Inhibitors on the Solvent Exposed Region: From Symmetric to Asymmetric Naphthalenesulfonamides.
J.Med.Chem., 65, 2022
|
|
7XM5
 
 | | Keap1 Kelch domain (residues 322-609) in complex with 6i | | Descriptor: | Kelch-like ECH-associated protein 1, N-[4-[(2-azanyl-2-oxidanylidene-ethyl)-[4-[(2-azanyl-2-oxidanylidene-ethyl)-(4-methoxyphenyl)sulfonyl-amino]naphthalen-1-yl]sulfamoyl]phenyl]-3-morpholin-4-yl-propanamide | | Authors: | Xu, K. | | Deposit date: | 2022-04-24 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallography-Guided Optimizations of the Keap1-Nrf2 Inhibitors on the Solvent Exposed Region: From Symmetric to Asymmetric Naphthalenesulfonamides.
J.Med.Chem., 65, 2022
|
|
2H96
 
 | | Discovery of Potent, Highly Selective, and Orally Bioavailable Pyridine Carboxamide C-jun NH2-terminal Kinase Inhibitors | | Descriptor: | 5-CYANO-N-(2,5-DIMETHOXYBENZYL)-6-ETHOXYPYRIDINE-2-CARBOXAMIDE, C-jun-amino-terminal kinase-interacting protein 1, GLYCEROL, ... | | Authors: | Abad-Zapatero, C. | | Deposit date: | 2006-06-09 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of potent, highly selective, and orally bioavailable pyridine carboxamide c-Jun NH2-terminal kinase inhibitors.
J.Med.Chem., 49, 2006
|
|
4YNE
 
 | | (R)-2-Phenylpyrrolidine Substitute Imidazopyridazines: a New Class of Potent and Selective Pan-TRK Inhibitors | | Descriptor: | 6-[(2R)-2-(3-fluorophenyl)pyrrolidin-1-yl]-3-(pyridin-2-yl)imidazo[1,2-b]pyridazine, GLYCEROL, High affinity nerve growth factor receptor, ... | | Authors: | Kreusch, A, Rucker, P, Molteni, V, Loren, J. | | Deposit date: | 2015-03-09 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.0229 Å) | | Cite: | (R)-2-Phenylpyrrolidine Substituted Imidazopyridazines: A New Class of Potent and Selective Pan-TRK Inhibitors.
Acs Med.Chem.Lett., 6, 2015
|
|
