7EW8
 
 | | Legionella pneumophila effector AnkD | | Descriptor: | ANK_REP_REGION domain-containing protein | | Authors: | Chen, T.T, Lin, Y.L. | | Deposit date: | 2021-05-24 | | Release date: | 2022-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Atypical Legionella GTPase effector hijacks host vesicular transport factor p115 to regulate host lipid droplet.
Sci Adv, 8, 2022
|
|
4M6V
 
 | |
4LOC
 
 | |
6LAN
 
 | | Structure of CCDC50 and LC3B complex | | Descriptor: | Coiled-coil domain-containing protein 50,Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Liu, L, Li, J, Hou, P. | | Deposit date: | 2019-11-12 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection.
Cell Res., 31, 2021
|
|
5XOB
 
 | | Crystal structure of apo TiaS (tRNAIle2 agmatidine synthetase) | | Descriptor: | MAGNESIUM ION, ZINC ION, tRNA(Ile2) 2-agmatinylcytidine synthetase TiaS | | Authors: | Dong, J. | | Deposit date: | 2017-05-27 | | Release date: | 2018-08-29 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of tRNA-Modifying Enzyme TiaS and Motions of Its Substrate Binding Zinc Ribbon.
J. Mol. Biol., 430, 2018
|
|
6AGG
 
 | |
7D8R
 
 | | MITF HLHLZ structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8T
 
 | | MITF bHLHLZ complex with M-box DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*AP*CP*AP*TP*GP*TP*GP*TP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*AP*CP*AP*CP*AP*TP*GP*TP*TP*AP*CP*AP*G)-3'), Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7D8S
 
 | | MITF bHLHLZ apo structure | | Descriptor: | Microphthalmia-associated transcription factor,Methionyl-tRNA synthetase beta subunit, SULFATE ION | | Authors: | Guo, M, Fang, P, Wang, J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7EOD
 
 | | MITF HLHLZ Delta AKE | | Descriptor: | GLYCEROL, Isoform M1 of Microphthalmia-associated transcription factor | | Authors: | Li, P, Liu, Z, Fang, P, Wang, J. | | Deposit date: | 2021-04-22 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Cell Res., 33, 2023
|
|
7WM0
 
 | |
7WLY
 
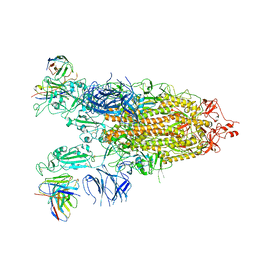 | | Cryo-EM structure of the Omicron S in complex with 35B5 Fab(1 down- and 2 up RBDs) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, ... | | Authors: | Wang, X, Zhu, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | 35B5 antibody potently neutralizes SARS-CoV-2 Omicron by disrupting the N-glycan switch via a conserved spike epitope.
Cell Host Microbe, 30, 2022
|
|
7WLZ
 
 | |
6B1B
 
 | | STRUCTURE OF 4-HYDROXYPHENYLACETATE 3-MONOOXYGENASE (HPAB), OXYGENASE COMPONENT FROM ESCHERICHIA COLI MUTANT XS6 (APO Enzyme) | | Descriptor: | 4-hydroxyphenylacetate 3-monooxygenase, oxygenase subunit, trimethylamine oxide | | Authors: | Zhou, D, Kandavelu, P, Wang, B.C, Yan, Y, Rose, J.P. | | Deposit date: | 2017-09-18 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Structural Insights into Catalytic Versatility of the Flavin-dependent Hydroxylase (HpaB) from Escherichia coli.
Sci Rep, 9, 2019
|
|
5Y9O
 
 | | The apo structure of Legionella pneumophila WipA | | Descriptor: | IODIDE ION, WipA | | Authors: | Xie, W, Jia, Q. | | Deposit date: | 2017-08-26 | | Release date: | 2018-05-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Legionella pneumophila effector WipA, a bacterial PPP protein phosphatase with PTP activity
Acta Biochim. Biophys. Sin. (Shanghai), 50, 2018
|
|
8IBY
 
 | | Structure of R2 with 5'ORF | | Descriptor: | 5'ORF RNA, Reverse transcriptase-like protein, ZINC ION | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBX
 
 | | Structure of R2 with 3'UTR and DNA in unwinding state | | Descriptor: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBW
 
 | | Structure of R2 with 3'UTR and DNA in binding state | | Descriptor: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBZ
 
 | | Structure of R2 with 5'ORF and 3'UTR | | Descriptor: | 5ORF-linker-3UTR, Reverse transcriptase-like protein, ZINC ION | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
2QYF
 
 | |
1KC4
 
 | | NMR Structural Analysis of the Complex Formed Between alpha-Bungarotoxin and the Principal alpha-Neurotoxin Binding Sequence on the alpha7 Subunit of a Neuronal Nicotinic Acetylcholine Receptor | | Descriptor: | alpha-bungarotoxin, neuronal acetylcholine receptor protein, alpha-7 chain | | Authors: | Moise, L, Piserchio, A, Basus, V.J, Hawrot, E. | | Deposit date: | 2001-11-07 | | Release date: | 2002-03-13 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of alpha-bungarotoxin and its complex with the principal alpha-neurotoxin-binding sequence on the alpha 7 subunit of a neuronal nicotinic acetylcholine receptor.
J.Biol.Chem., 277, 2002
|
|
1KL8
 
 | | NMR STRUCTURAL ANALYSIS OF THE COMPLEX FORMED BETWEEN ALPHA-BUNGAROTOXIN AND THE PRINCIPAL ALPHA-NEUROTOXIN BINDING SEQUENCE ON THE ALPHA7 SUBUNIT OF A NEURONAL NICOTINIC ACETYLCHOLINE RECEPTOR | | Descriptor: | ALPHA-BUNGAROTOXIN, NEURONAL ACETYLCHOLINE RECEPTOR PROTEIN, ALPHA-7 CHAIN | | Authors: | Moise, L, Piserchio, A, Basus, V.J, Hawrot, E. | | Deposit date: | 2001-12-11 | | Release date: | 2002-03-13 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of alpha-bungarotoxin and its complex with the principal alpha-neurotoxin-binding sequence on the alpha 7 subunit of a neuronal nicotinic acetylcholine receptor.
J.Biol.Chem., 277, 2002
|
|
7WCN
 
 | | Cryo-EM structure of GPR119-Gs Complex with small molecule agonist AR231453 | | Descriptor: | Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qiao, A.N, Wu, S, Ye, S. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Activation and signaling mechanism revealed by GPR119-G s complex structures.
Nat Commun, 13, 2022
|
|
7WCM
 
 | | Cryo-EM structure of GPR119-Gs Complex with small molecule agonist MBX-2982 | | Descriptor: | 2-[1-(5-ethylpyrimidin-2-yl)piperidin-4-yl]-4-[[4-(1,2,3,4-tetrazol-1-yl)phenoxy]methyl]-1,3-thiazole, Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qiao, A.N, Wu, S, Ye, S. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Activation and signaling mechanism revealed by GPR119-G s complex structures.
Nat Commun, 13, 2022
|
|
1S2H
 
 | | The Mad2 spindle checkpoint protein possesses two distinct natively folded states | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2A | | Authors: | Luo, X, Tang, Z, Xia, G, Wassmann, K, Matsumoto, T, Rizo, J, Yu, H. | | Deposit date: | 2004-01-08 | | Release date: | 2004-03-30 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | The Mad2 spindle checkpoint protein has two distinct natively folded states.
Nat.Struct.Mol.Biol., 11, 2004
|
|
