4IVA
 
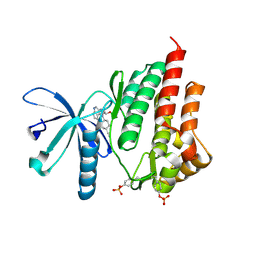 | | JAK2 kinase (JH1 domain) in complex with the inhibitor TRANS-4-[(8AS)-2-[(1R)-1-HYDROXYETHYL]IMIDAZO[4,5-D]PYRROLO[2,3-B]PYRIDIN-1(8AH)-YL]CYCLOHEXANECARBONITRILE | | Descriptor: | Tyrosine-protein kinase JAK2, trans-4-{2-[(1R)-1-hydroxyethyl]imidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(6H)-yl}cyclohexanecarbonitrile | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2013-01-22 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of C-2 Hydroxyethyl Imidazopyrrolopyridines as Potent JAK1 Inhibitors with Favorable Physicochemical Properties and High Selectivity over JAK2.
J.Med.Chem., 56, 2013
|
|
6AXQ
 
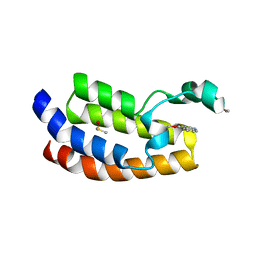 | |
6AY5
 
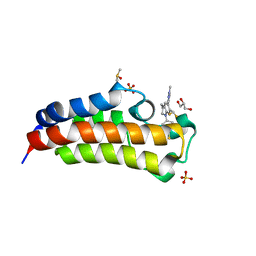 | | CREBBP bromodomain in complex with Cpd17 (5-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-3-methylbenzo[d]thiazol-2(3H)-one) | | Descriptor: | 5-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-3-methyl-1,3-benzothiazol-2(3H)-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A Unique Approach to Design Potent and Selective Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP) Inhibitors.
J. Med. Chem., 60, 2017
|
|
6AY3
 
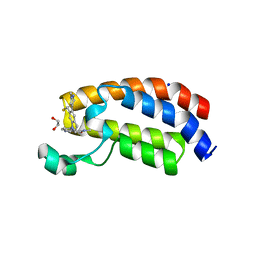 | | CREBBP bromodomain in complex with Cpd16 (5-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-N-methyl-1H-indole-3-carboxamide) | | Descriptor: | 1,2-ETHANEDIOL, 5-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-N-methyl-1H-indole-3-carboxamide, CREB-binding protein, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | A Unique Approach to Design Potent and Selective Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP) Inhibitors.
J. Med. Chem., 60, 2017
|
|
4F08
 
 | |
4F09
 
 | |
6ALC
 
 | | CREBBP bromodomain in complex with Cpd 4 (1-(1-(cyclopropylmethyl)-3-(1H-indol-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1,2-ETHANEDIOL, 1-[1-(cyclopropylmethyl)-3-(1H-indol-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl]ethan-1-one, CREB-binding protein, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-08-07 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | Design and synthesis of a biaryl series as inhibitors for the bromodomains of CBP/P300.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
4EHZ
 
 | | The Jak1 kinase domain in complex with inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-1-(piperidin-4-yl)-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridine, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Steffek, M. | | Deposit date: | 2012-04-04 | | Release date: | 2012-07-04 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Discovery and optimization of C-2 methyl imidazopyrrolopyridines as potent and orally bioavailable JAK1 inhibitors with selectivity over JAK2.
J.Med.Chem., 55, 2012
|
|
4EI4
 
 | | JAK1 kinase (JH1 domain) in complex with compound 20 | | Descriptor: | (1R,3R)-3-(2-methylimidazo[4,5-d]pyrrolo[2,3-b]pyridin-1(8H)-yl)cyclohexanol, Tyrosine-protein kinase JAK1 | | Authors: | Eigenbrot, C, Steffek, M. | | Deposit date: | 2012-04-04 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery and optimization of C-2 methyl imidazopyrrolopyridines as potent and orally bioavailable JAK1 inhibitors with selectivity over JAK2.
J.Med.Chem., 55, 2012
|
|
1JYB
 
 | | Crystal structure of Rubrerythrin | | Descriptor: | FE (III) ION, Rubrerythrin, ZINC ION | | Authors: | Chang, W.R, Li, M, Liu, M.Y. | | Deposit date: | 2001-09-11 | | Release date: | 2002-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure studies on rubrerythrin: enzymatic activity in relation to the zinc movement.
J.Biol.Inorg.Chem., 8, 2003
|
|
5V9P
 
 | | Crystal structure of pyrrolidine amide inhibitor [(3S)-3-(4-bromo-1H-pyrazol-1-yl)pyrrolidin-1-yl][3-(propan-2-yl)-1H-pyrazol-5-yl]methanone (compound 35) in complex with KDM5A | | Descriptor: | Lysine-specific demethylase 5A, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Kiefer, J.R, Liang, J, Vinogradova, M. | | Deposit date: | 2017-03-23 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | From a novel HTS hit to potent, selective, and orally bioavailable KDM5 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5V9T
 
 | | Crystal structure of selective pyrrolidine amide KDM5a inhibitor N-{(3R)-1-[3-(propan-2-yl)-1H-pyrazole-5-carbonyl]pyrrolidin-3-yl}cyclopropanecarboxamide (compound 48) | | Descriptor: | Lysine-specific demethylase 5A, N-{(3R)-1-[3-(propan-2-yl)-1H-pyrazole-5-carbonyl]pyrrolidin-3-yl}cyclopropanecarboxamide, NICKEL (II) ION, ... | | Authors: | Kiefer, J.R, Liang, J, Vinogradova, M. | | Deposit date: | 2017-03-23 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | From a novel HTS hit to potent, selective, and orally bioavailable KDM5 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
8HKD
 
 | |
6LK5
 
 | | MLKL mutant - T357ES358D | | Descriptor: | Mixed lineage kinase domain-like protein | | Authors: | Wang, H.Y, Li, S, Zhang, Y. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The MLKL kinase-like domain dimerization is an indispensable step of mammalian MLKL activation in necroptosis signaling.
Cell Death Dis, 12, 2021
|
|
6LK6
 
 | | MLKL mutant - T357AS358A | | Descriptor: | Mixed lineage kinase domain-like protein | | Authors: | Wang, H, Li, S, Zhang, Y. | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The MLKL kinase-like domain dimerization is an indispensable step of mammalian MLKL activation in necroptosis signaling.
Cell Death Dis, 12, 2021
|
|
1JDQ
 
 | | Solution Structure of TM006 Protein from Thermotoga maritima | | Descriptor: | HYPOTHETICAL PROTEIN TM0983 | | Authors: | Denisov, A.Y, Finak, G, Yee, A, Kozlov, G, Gehring, K, Arrowsmith, C.H. | | Deposit date: | 2001-06-14 | | Release date: | 2002-02-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JCU
 
 | |
1JRM
 
 | |
1JE3
 
 | | Solution Structure of EC005 from Escherichia coli | | Descriptor: | HYPOTHETICAL 8.6 KDA PROTEIN IN AMYA-FLIE INTERGENIC REGION | | Authors: | Yee, A, Gutierrez, P, Kozlov, G, Denisov, A, Gehring, K, Arrowsmith, C. | | Deposit date: | 2001-06-15 | | Release date: | 2002-03-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JW2
 
 | | SOLUTION STRUCTURE OF HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha FROM ESCHERICHIA COLI. Ontario Centre for Structural Proteomics target EC0308_1_72; Northeast Structural Genomics Target ET88 | | Descriptor: | HEMOLYSIN EXPRESSION MODULATING PROTEIN Hha | | Authors: | Chang, X, Yee, A, Savchenko, A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JW3
 
 | | Solution Structure of Methanobacterium Thermoautotrophicum Protein 1598. Ontario Centre for Structural Proteomics target MTH1598_1_140; Northeast Structural Genomics Target TT6 | | Descriptor: | Conserved Hypothetical Protein MTH1598 | | Authors: | Chang, X, Connelly, G, Yee, A, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-09-02 | | Release date: | 2002-02-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
5WYM
 
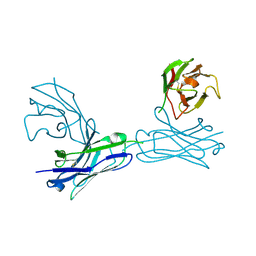 | | Crystal structure of an anti-connexin26 scFv | | Descriptor: | anti-connexin26 scFv,Ig heavy chain,Linker,anti-connexin26 scFv,Ig light chain | | Authors: | Li, S, Xu, L. | | Deposit date: | 2017-01-13 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Design and Characterization of a Human Monoclonal Antibody that Modulates Mutant Connexin 26 Hemichannels Implicated in Deafness and Skin Disorders
Front Mol Neurosci, 10, 2017
|
|
5B30
 
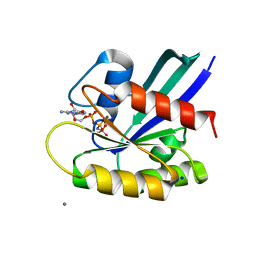 | | H-Ras WT in complex with GppNHp (state 1) after structural transition by humidity control | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Kumasaka, T, Miyano, N, Baba, S, Matsumoto, S, Kataoka, T, Shima, F. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Mechanism for Conformational Dynamics of Ras-GTP Elucidated from In-Situ Structural Transition in Crystal
Sci Rep, 6, 2016
|
|
5B2Z
 
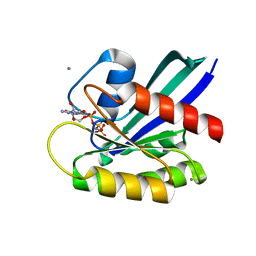 | | H-Ras WT in complex with GppNHp (state 2*) before structural transition by humidity control | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Kumasaka, T, Miyano, N, Baba, S, Matsumoto, S, Kataoka, T, Shima, F. | | Deposit date: | 2016-02-07 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Molecular Mechanism for Conformational Dynamics of Ras-GTP Elucidated from In-Situ Structural Transition in Crystal
Sci Rep, 6, 2016
|
|
5CET
 
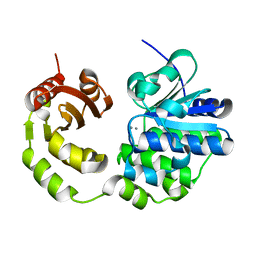 | | Crystal structure of Rv2837c | | Descriptor: | Bifunctional oligoribonuclease and PAP phosphatase NrnA, MANGANESE (II) ION | | Authors: | Wang, F, He, Q, Zhu, D, Liu, S, Gu, L. | | Deposit date: | 2015-07-07 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Biochemical Insight into the Mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP Phosphodiesterase
J.Biol.Chem., 291, 2016
|
|
