7XKJ
 
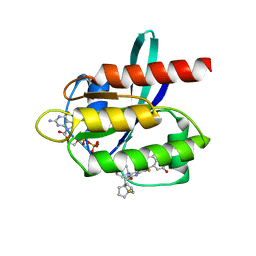 | | Kras-G12D-GDP-MRTX1133 by FIB-MicroED | | Descriptor: | 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)-5-ethynyl-6-fluoronaphthalen-2-ol, GUANOSINE-5'-DIPHOSPHATE, KRAS proto-oncogene, ... | | Authors: | Li, X.M. | | Deposit date: | 2022-04-19 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (3 Å) | | Cite: | Kras-G12D-GDP-MRTX1133 by FIB-MicroED
To Be Published
|
|
9K9U
 
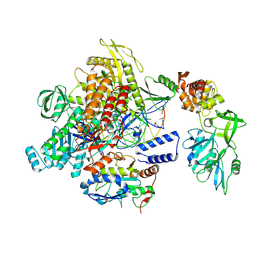 | | MPXV DNA polymerase complex in editing state 1 | | Descriptor: | DNA (25-MER), DNA (38-MER), DNA polymerase, ... | | Authors: | Xie, Y.F, Kuai, L, Peng, Q, Wang, Q, Wang, H, Li, X.M, Qi, J.X, Ding, Q, Shi, Y, Gao, F. | | Deposit date: | 2024-10-27 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | MPXV DNA polymerase complex in editing state 1
To Be Published
|
|
9K9S
 
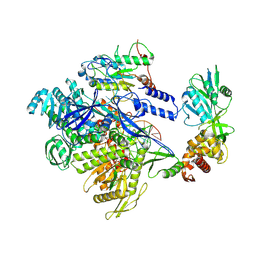 | | MPXV DNA polymerase in complex with primer/4U template DNA | | Descriptor: | DNA (25-MER), DNA (4U 38-MER), DNA polymerase, ... | | Authors: | Xie, Y.F, Kuai, L, Peng, Q, Wang, Q, Wang, H, Li, X.M, Qi, J.X, Ding, Q, Shi, Y, Gao, F. | | Deposit date: | 2024-10-27 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | MPXV DNA polymerase in complex with primer/4U template DNA
To Be Published
|
|
9K9V
 
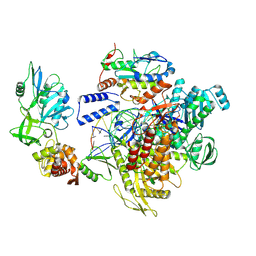 | | MPXV DNA polymerase complex in editing state 2 | | Descriptor: | DNA (25-MER), DNA (38-MER), DNA polymerase, ... | | Authors: | Xie, Y.F, Kuai, L, Peng, Q, Wang, Q, Wang, H, Li, X.M, Qi, J.X, Ding, Q, Shi, Y, Gao, F. | | Deposit date: | 2024-10-27 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | MPXV DNA polymerase complex in editing state 2
To Be Published
|
|
9K9R
 
 | | MPXV DNA polymerase in complex with primer/5U template DNA | | Descriptor: | DNA (25-MER), DNA (5U 38-MER), DNA polymerase, ... | | Authors: | Xie, Y.F, Kuai, L, Peng, Q, Wang, Q, Wang, H, Li, X.M, Qi, J.X, Ding, Q, Shi, Y, Gao, F. | | Deposit date: | 2024-10-27 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | MPXV DNA polymerase in complex with primer/5U template DNA
To Be Published
|
|
9K9T
 
 | | MPXV DNA polymerase in complex with CDV | | Descriptor: | DNA (24-MER), DNA (38-MER), DNA polymerase, ... | | Authors: | Xie, Y.F, Kuai, L, Peng, Q, Wang, Q, Wang, H, Li, X.M, Qi, J.X, Ding, Q, Shi, Y, Gao, F. | | Deposit date: | 2024-10-27 | | Release date: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | MPXV DNA polymerase in complex with CDV
To Be Published
|
|
3MCB
 
 | | Crystal structure of NAC domains of human nascent polypeptide-associated complex (NAC) | | Descriptor: | IODIDE ION, Nascent polypeptide-associated complex subunit alpha, Transcription factor BTF3 | | Authors: | Wang, L.F, Zhang, W.C, Wang, L, Zhang, X.J.C, Li, X.M, Rao, Z. | | Deposit date: | 2010-03-29 | | Release date: | 2010-07-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of NAC domains of human nascent polypeptide-associated complex (NAC) and its alphaNAC subunit
Protein Cell, 1, 2010
|
|
3MCE
 
 | | Crystal structure of the NAC domain of alpha subunit of nascent polypeptide-associated complex(NAC) | | Descriptor: | IODIDE ION, Nascent polypeptide-associated complex subunit alpha | | Authors: | Wang, L.F, Zhang, W.C, Wang, L, Zhang, X.J.C, Li, X.M, Rao, Z. | | Deposit date: | 2010-03-29 | | Release date: | 2010-07-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Crystal structures of NAC domains of human nascent polypeptide-associated complex (NAC) and its alphaNAC subunit
Protein Cell, 1, 2010
|
|
4PR3
 
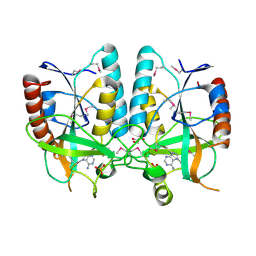 | | Crystal structure of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Descriptor: | 5'-methylthioadenosine nucleosidase / s-adenosylhomocysteine nucleosidase, ADENINE, GLYCEROL, ... | | Authors: | Zhang, X.C, Kang, X.S, Zhao, Y, Jiang, D.H, Li, X.M, Chen, Z.L. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Crystal structure and biochemical studies of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Biochem.Biophys.Res.Commun., 446, 2014
|
|
7CQ7
 
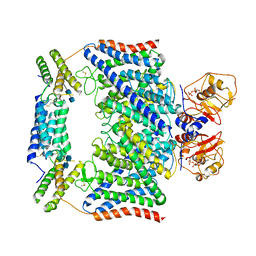 | | Structure of the human CLCN7-OSTM1 complex with ADP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2020-08-08 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structure of the human CLCN7-OSTM1 complex with ADP
To Be Published
|
|
7CQ5
 
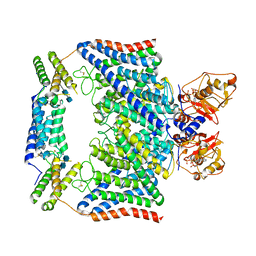 | | Structure of the human CLCN7-OSTM1 complex with ATP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2020-08-08 | | Release date: | 2020-11-11 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of the human CLCN7-OSTM1 complex with ATP
To Be Published
|
|
7CQ6
 
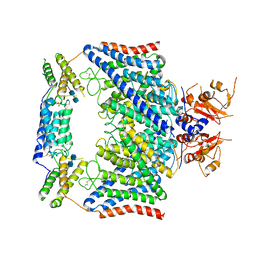 | | Structure of the human CLCN7-OSTM1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, H(+)/Cl(-) exchange transporter 7, ... | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2020-08-08 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the human CLCN7-OSTM1 complex
To Be Published
|
|
3K2S
 
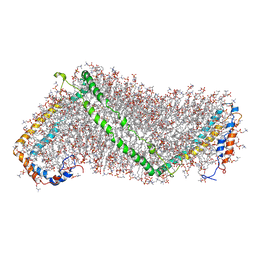 | | Solution structure of double super helix model | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Apolipoprotein A-I, CHOLESTEROL | | Authors: | Wu, Z, Gogonea, V, Lee, X, Wagner, M.A, Li, X.-M, Huang, Y, Undurti, A, May, R.P, Haertlein, M, Moulin, M, Gutsche, I, Zaccai, G, Didonato, J.A, Hazen, L.S. | | Deposit date: | 2009-09-30 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Double superhelix model of high density lipoprotein.
J.Biol.Chem., 284, 2009
|
|
5YYF
 
 | |
3MTV
 
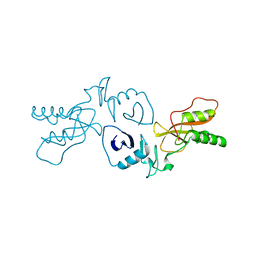 | | The Crystal Structure of the PRRSV Nonstructural Protein Nsp1 | | Descriptor: | Papain-like cysteine protease | | Authors: | Xue, F, Sun, Y.N, Yan, L.M, Zhao, C, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2010-04-30 | | Release date: | 2010-05-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of porcine reproductive and respiratory syndrome virus nonstructural protein Nsp1beta reveals a novel metal-dependent nuclease
J.Virol., 84, 2010
|
|
3NX2
 
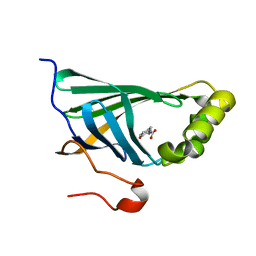 | | Enterobacter sp. Px6-4 Ferulic Acid Decarboxylase in complex with substrate analogues | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Ferulic acid decarboxylase | | Authors: | Gu, W, Yang, J.K, Lou, Z.Y, Meng, Z.H, Zhang, K.-Q. | | Deposit date: | 2010-07-12 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis of Enzymatic Activity for the Ferulic Acid Decarboxylase (FADase) from Enterobacter sp. Px6-4
Plos One, 6, 2011
|
|
3NX1
 
 | | Crystal structure of Enterobacter sp. Px6-4 Ferulic Acid Decarboxylase | | Descriptor: | Ferulic acid decarboxylase | | Authors: | Gu, W, Yang, J.K, Lou, Z.Y, Meng, Z.H, Zhang, K.-Q. | | Deposit date: | 2010-07-12 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Enzymatic Activity for the Ferulic Acid Decarboxylase (FADase) from Enterobacter sp. Px6-4
Plos One, 6, 2011
|
|
8WU1
 
 | | Cryo-EM structure of CB1-beta-arrestin1 complex | | Descriptor: | Beta-arrestin-1, Cannabinoid receptor 1,Vasopressin V2 receptor, Fab30 heavy chain, ... | | Authors: | Liao, Y, Zhang, H, Shen, Q, Cai, C. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Snapshot of the cannabinoid receptor 1-arrestin complex unravels the biased signaling mechanism.
Cell, 186, 2023
|
|
8IA7
 
 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | CCK-8, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2023-02-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
6L5Z
 
 | |
6ISO
 
 | | Human SIRT3 Recognizing H3K4cr | | Descriptor: | (2E)-BUT-2-ENAL, ARG-THR-LYS-GLN-THR-ALA-ARG, GLYCEROL, ... | | Authors: | Wang, Y, Hao, Q. | | Deposit date: | 2018-11-17 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Identification of 'erasers' for lysine crotonylated histone marks using a chemical proteomics approach.
Elife, 3, 2014
|
|
7XOU
 
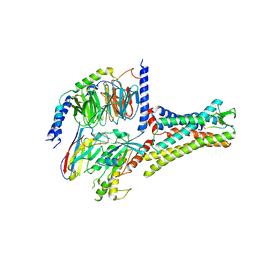 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | CCK-8, Cholecystokinin receptor type A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
7XOW
 
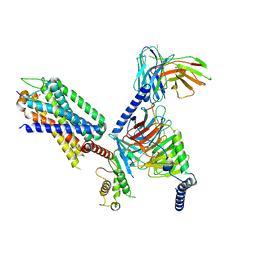 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | Gastrin, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
7XOV
 
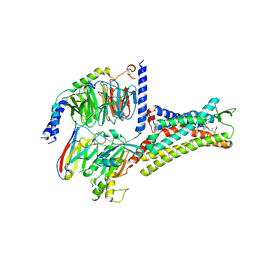 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | 2-[2-[[4-(4-chloranyl-2,5-dimethoxy-phenyl)-5-(2-cyclohexylethyl)-1,3-thiazol-2-yl]carbamoyl]-5,7-dimethyl-indol-1-yl]ethanoic acid, Cholecystokinin receptor type A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
7WSW
 
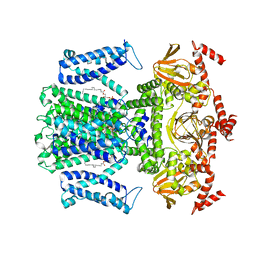 | | Cryo-EM structure of the Potassium channel AKT1 from Arabidopsis thaliana | | Descriptor: | PHOSPHATIDYLETHANOLAMINE, POTASSIUM ION, Potassium channel AKT1 | | Authors: | Yang, G.H, Lu, Y.M, Zhang, Y.M, Jia, Y.T, Li, X.M, Lei, J.L. | | Deposit date: | 2022-02-02 | | Release date: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis.
Nat Commun, 13, 2022
|
|
