8AD9
 
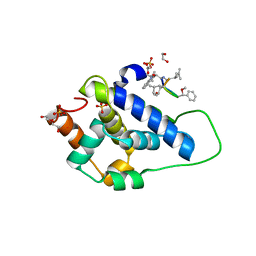 | | Crystal structure of ClpC2 C-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cyclomarin A, ... | | Authors: | Taylor, G, Cui, H.J, Leodolter, J, Giese, C, Weber-Ban, E. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | ClpC2 protects mycobacteria against a natural antibiotic targeting ClpC1-dependent protein degradation.
Commun Biol, 6, 2023
|
|
8ADA
 
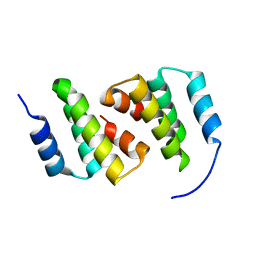 | | Crystal structure of ClpC2 N-terminal domain | | Descriptor: | Uncharacterized protein Rv2667 | | Authors: | Taylor, G, Cui, H.J, Leodolter, J, Giese, C, Weber-Ban, E. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | ClpC2 protects mycobacteria against a natural antibiotic targeting ClpC1-dependent protein degradation.
Commun Biol, 6, 2023
|
|
6TV6
 
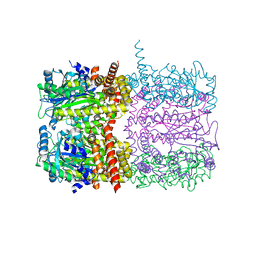 | | Octameric McsB from Bacillus subtilis. | | Descriptor: | MAGNESIUM ION, Protein-arginine kinase | | Authors: | Suskiewicz, M.J, Hajdusits, B, Meinhart, A, Clausen, T. | | Deposit date: | 2020-01-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | McsB forms a gated kinase chamber to mark aberrant bacterial proteins for degradation.
Elife, 10, 2021
|
|
7AA4
 
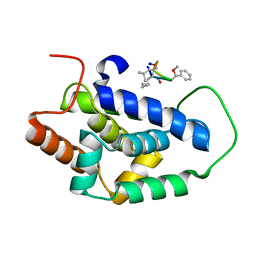 | | Structure of ClpC1-NTD bound to a CymA analogue | | Descriptor: | Negative regulator of genetic competence ClpC/mecB, polymer Cyclomarin A analogue | | Authors: | Meinhart, A, Morreale, F.E, Kaiser, M, Clausen, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
7ABR
 
 | | Cryo-EM structure of B. subtilis ClpC (DWB mutant) hexamer bound to a substrate polypeptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Negative regulator of genetic competence ClpC/MecB, ... | | Authors: | Morreale, F.E, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2020-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
8B9O
 
 | |
8B9U
 
 | |
