2VGG
 
 | | HUMAN ERYTHROCYTE PYRUVATE KINASE: R479H MUTANT | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Valentini, G, Chiarelli, L.R, Fortin, R, Dolzan, M, Galizzi, A, Abraham, D.J, Wang, C, Bianchi, P, Zanella, A, Mattevi, A. | | Deposit date: | 2007-11-13 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure and Function of Human Erythrocyte Pyruvate Kinase. Molecular Basis of Nonspherocytic Hemolytic Anemia.
J.Biol.Chem., 277, 2002
|
|
2VGF
 
 | | HUMAN ERYTHROCYTE PYRUVATE KINASE: T384M mutant | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Valentini, G, Chiarelli, L.R, Fortin, R, Dolzan, M, Galizzi, A, Abraham, D.J, Wang, C, Bianchi, P, Zanella, A, Mattevi, A. | | Deposit date: | 2007-11-12 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and Function of Human Erythrocyte Pyruvate Kinase. Molecular Basis of Nonspherocytic Hemolytic Anemia.
J.Biol.Chem., 277, 2002
|
|
2VGB
 
 | | HUMAN ERYTHROCYTE PYRUVATE KINASE | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Valentini, G, Chiarelli, L, Fortin, R, Dolzan, M, Galizzi, A, Abraham, D.J, Wang, C, Bianchi, P, Zanella, A, Mattevi, A. | | Deposit date: | 2007-11-12 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure and Function of Human Erythrocyte Pyruvate Kinase. Molecular Basis of Nonspherocytic Hemolytic Anemia.
J.Biol.Chem., 277, 2002
|
|
2VGI
 
 | | HUMAN ERYTHROCYTE PYRUVATE KINASE: R486W MUTANT | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Valentini, G, Chiarelli, L, Fortin, R, Dolzan, M, Galizzi, A, Abraham, D.J, Wang, C, Bianchi, P, Zanella, A, Mattevi, A. | | Deposit date: | 2007-11-13 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structure and Function of Human Erythrocyte Pyruvate Kinase. Molecular Basis of Nonspherocytic Hemolytic Anemia.
J.Biol.Chem., 277, 2002
|
|
3CZ4
 
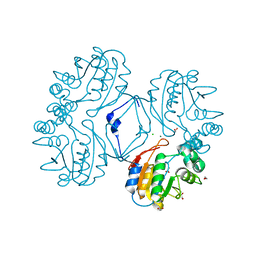 | | Native AphA class B acid phosphatase/phosphotransferase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Leone, R, Cappelletti, E, Benvenuti, M, Lentini, G, Thaller, M.C, Mangani, S. | | Deposit date: | 2008-04-28 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic mechanism of the bacterial class B phosphatase AphA belonging to the DDDD superfamily of phosphohydrolases.
J.Mol.Biol., 384, 2008
|
|
1S19
 
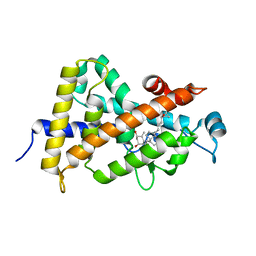 | | Crystal structure of VDR ligand binding domain complexed to calcipotriol. | | Descriptor: | CALCIPOTRIOL, Vitamin D3 receptor | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Moras, D. | | Deposit date: | 2004-01-06 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the vitamin D nuclear receptor liganded with the vitamin D side chain analogues calcipotriol and seocalcitol, receptor agonists of clinical importance. Insights into a structural basis for the switching of calcipotriol to a receptor antagonist by further side chain modification.
J.Med.Chem., 47, 2004
|
|
1IE8
 
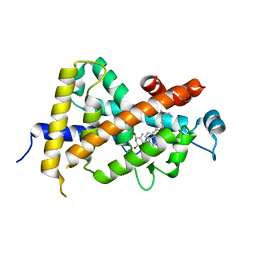 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to KH1060 | | Descriptor: | 5-(2-{1-[1-(4-ETHYL-4-HYDROXY-HEXYLOXY)-ETHYL]-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE}-ETHYLIDENE)-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1IE9
 
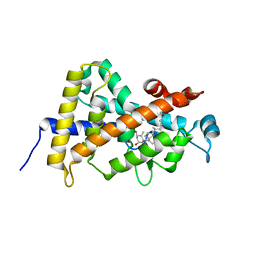 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to MC1288 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1S0Z
 
 | | Crystal structure of the VDR LBD complexed to seocalcitol. | | Descriptor: | SEOCALCITOL, Vitamin D3 receptor | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Moras, D. | | Deposit date: | 2004-01-05 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the vitamin D nuclear receptor liganded with the vitamin D side chain analogues calcipotriol and seocalcitol, receptor agonists of clinical importance. Insights into a structural basis for the switching of calcipotriol to a receptor antagonist by further side chain modification.
J.Med.Chem., 47, 2004
|
|
7NUR
 
 | |
1E0T
 
 | |
1E0U
 
 | |
1TXI
 
 | | Crystal structure of the vdr ligand binding domain complexed to TX522 | | Descriptor: | (1R,3R)-5-((Z)-2-((1R,7AS)-HEXAHYDRO-1-((S)-6-HYDROXY-6-METHYLHEPT-4-YN-2-YL)-7A-METHYL-1H-INDEN-4(7AH)-YLIDENE)ETHYLIDENE)CYCLOHEXANE-1,3-DIOL, Vitamin D receptor | | Authors: | Moras, D, Rochel, N. | | Deposit date: | 2004-07-05 | | Release date: | 2005-05-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Superagonistic Action of 14-epi-Analogs of 1,25-Dihydroxyvitamin D Explained by Vitamin D Receptor-Coactivator Interaction
Mol.Pharmacol., 67, 2005
|
|
1PKY
 
 | | PYRUVATE KINASE FROM E. COLI IN THE T-STATE | | Descriptor: | PYRUVATE KINASE | | Authors: | Mattevi, A. | | Deposit date: | 1995-04-27 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Escherichia coli pyruvate kinase type I: molecular basis of the allosteric transition.
Structure, 3, 1995
|
|
6R85
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-glutamate | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, Glutamate receptor 3.3,Glutamate receptor 3.3, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-03-31 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1FB5
 
 | | LOW RESOLUTION STRUCTURE OF OVINE ORNITHINE TRANSCARBMOYLASE IN THE UNLIGANDED STATE | | Descriptor: | NORVALINE, ORNITHINE TRANSCARBAMOYLASE | | Authors: | Zanotti, G, Battistutta, R, Panzalorto, M, Francescato, P, Bruno, G, De Gregorio, A. | | Deposit date: | 2000-07-14 | | Release date: | 2003-08-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Functional and structural characterization of ovine ornithine transcarbamoylase.
Org.Biomol.Chem., 1, 2003
|
|
6R8A
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-methionine | | Descriptor: | Glutamate receptor 3.3,Glutamate receptor 3.3, METHIONINE, SODIUM ION, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R89
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with L-cysteine | | Descriptor: | CHLORIDE ION, CYSTEINE, GLYCEROL, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6R88
 
 | | Structure of Arabidopsis thaliana GLR3.3 ligand-binding domain in complex with glycine | | Descriptor: | CHLORIDE ION, GLYCEROL, GLYCINE, ... | | Authors: | Alfieri, A, Pederzoli, R, Costa, A. | | Deposit date: | 2019-04-01 | | Release date: | 2020-01-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural bases for agonist diversity in anArabidopsis thalianaglutamate receptor-like channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
