4JJ3
 
 | |
4JJ0
 
 | | Crystal structure of MamP | | Descriptor: | GLYCEROL, HEME C, MamP | | Authors: | Siponen, M, Pignol, D, Arnoux, P. | | Deposit date: | 2013-03-07 | | Release date: | 2013-10-09 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into magnetochrome-mediated magnetite biomineralization.
Nature, 502, 2013
|
|
7AV4
 
 | | Dark state structure of the C432S mutant of Fatty Acid Photodecarboxylase (FAP) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | Authors: | Schlichting, I, Hartmann, E, Arnoux, P, Sorigue, D, Beisson, F. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRV
 
 | | Crystal structure of FAP after illumination at 100K | | Descriptor: | CARBON DIOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRU
 
 | | Crystal structure of FAP in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YS2
 
 | | Crystal structure of FAP R451A in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YRX
 
 | | Low-dose crystal structure of FAP at room temperature | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
6YS1
 
 | | Crystal structure of FAP R451K mutant in the dark at 100K | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid Photodecarboxylase, STEARIC ACID, ... | | Authors: | Sorigue, D, Gotthard, G, Blangy, S, Nurizzo, D, Royant, A, Beisson, F, Arnoux, P. | | Deposit date: | 2020-04-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
7Z6O
 
 | | X-Ray studies of Ku70/80 reveal the binding site for IP6 | | Descriptor: | DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Varela, P.F, Charbonnier, J.B. | | Deposit date: | 2022-03-14 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
7ZT6
 
 | |
7ZVT
 
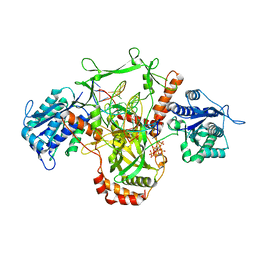 | | CryoEM structure of Ku heterodimer bound to DNA | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-17 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
7NPW
 
 | |
5OW4
 
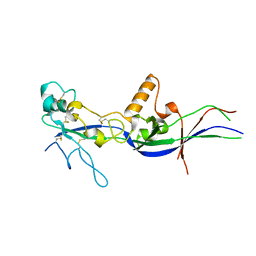 | |
7PU5
 
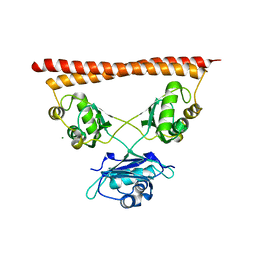 | | Structure of SFPQ-NONO complex | | Descriptor: | MAGNESIUM ION, Non-POU domain-containing octamer-binding protein, Splicing factor, ... | | Authors: | Fribourg, S. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Crystal structure of SFPQ-NONO heterodimer.
Biochimie, 198, 2022
|
|
7Q5A
 
 | | Lanreotide nanotube | | Descriptor: | Lanreotide | | Authors: | Pieri, L, Wang, F, Arteni, A.A, Bressanelli, S, Egelman, E.H, Paternostre, M. | | Deposit date: | 2021-11-03 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Atomic structure of Lanreotide nanotubes revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2X6S
 
 | | Human foamy virus integrase - catalytic core. Magnesium-bound structure. | | Descriptor: | INTEGRASE, MAGNESIUM ION | | Authors: | Rety, S, Delelis, O, Rezabkova, L, Dubanchet, B, Silhan, J, Lewit-Bentley, A. | | Deposit date: | 2010-02-19 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Studies of the Catalytic Core of the Primate Foamy Virus (Pfv-1) Integrase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2X6N
 
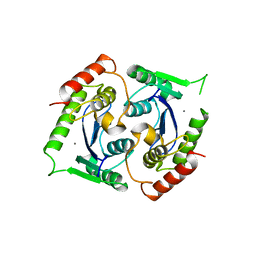 | | Human foamy virus integrase - catalytic core. Manganese-bound structure. | | Descriptor: | INTEGRASE, MANGANESE (II) ION | | Authors: | Rety, S, Delelis, O, Rezabkova, L, Dubanchet, B, Silhan, J, Lewit-Bentley, A. | | Deposit date: | 2010-02-18 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Studies of the Catalytic Core of the Primate Foamy Virus (Pfv-1) Integrase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5M2W
 
 | | Structure of nanobody nb18 raised against TssK from E. coli T6SS | | Descriptor: | Llama nanobody nb8 against TssK from T6SS, SULFATE ION | | Authors: | Cambillau, C, Nguyen, V.S, Spinelli, S, Desmyter, A. | | Deposit date: | 2016-10-13 | | Release date: | 2017-06-28 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
8CKE
 
 | |
8CH2
 
 | | PBP AccA from A. vitis S4 in complex with L-arabinose-2-phosphate (A2P) | | Descriptor: | 1,2-ETHANEDIOL, 2-O-phosphono-alpha-L-arabinopyranose, 2-O-phosphono-beta-L-arabinopyranose, ... | | Authors: | Morera, S, Deicsics, G, Vigouroux, A. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CKD
 
 | | PBP AccA from A. fabrum C58 in complex with agrocinopine D-like | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, Agrocinopine D-like (C2-C2 linked; with an alpha and beta-D-glucopyranose), ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-15 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CH1
 
 | | PBP AccA from A. vitis S4 in complex with Agrocinopine A | | Descriptor: | 1,2-ETHANEDIOL, 2-O-phosphono-alpha-L-arabinopyranose, 2-O-phosphono-beta-L-arabinopyranose, ... | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CJU
 
 | | PBP AccA from A. vitis S4 in complex with agrocin84 | | Descriptor: | ABC transporter substrate binding protein (Agrocinopine), [(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl]oxy-N-[9-[(2R,3S,5R)-5-[[[(2R,3S)-4-methyl-2,3-bis(oxidanyl)pentanoyl]amino]-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxolan-2-yl]purin-6-yl]phosphonamidic acid | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CAY
 
 | | PBP AccA from A. tumefaciens Bo542 in complex with Agrocinopine D-like | | Descriptor: | Agrocinopine D-like (C2-C2 linked; with an alpha and beta-D-glucopyranose), Agrocinopine D-like (C2-C2 linked; with two alpha-D-glucopyranoses), Agrocinopine utilization periplasmic binding protein AccA, ... | | Authors: | Morera, S, Vigouroux, A, Siragu, S. | | Deposit date: | 2023-01-24 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.626 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CH3
 
 | | PBP AccA from A. vitis S4 in complex with Agrocinopine C-like | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, Agrocinopine utilization periplasmic binding protein AccA, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
